Yunqing Liu
HD-Prot: A Protein Language Model for Joint Sequence-Structure Modeling with Continuous Structure Tokens
Dec 17, 2025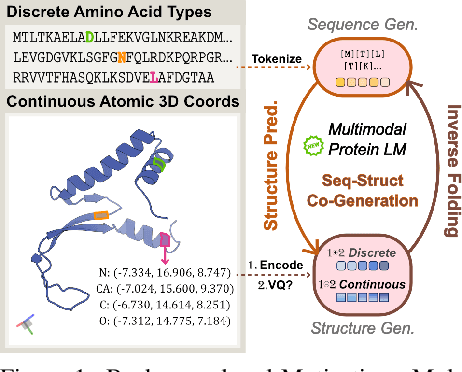
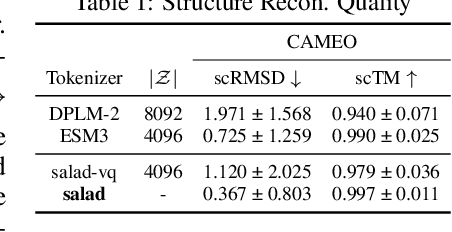
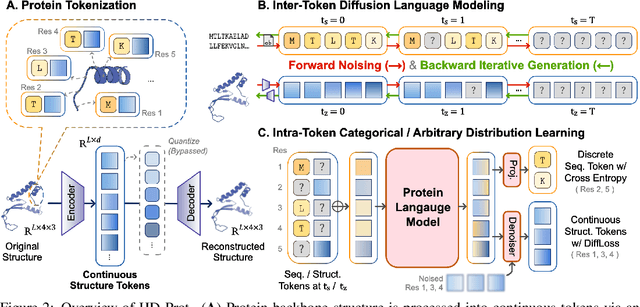
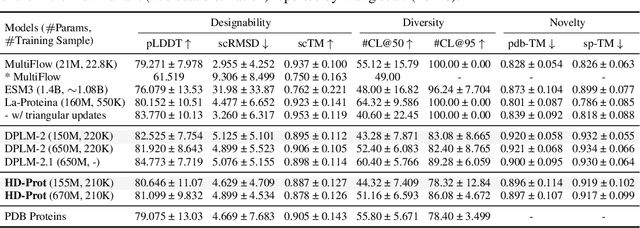
Abstract:Proteins inherently possess a consistent sequence-structure duality. The abundance of protein sequence data, which can be readily represented as discrete tokens, has driven fruitful developments in protein language models (pLMs). A key remaining challenge, however, is how to effectively integrate continuous structural knowledge into pLMs. Current methods often discretize protein structures to accommodate the language modeling framework, which inevitably results in the loss of fine-grained information and limits the performance potential of multimodal pLMs. In this paper, we argue that such concerns can be circumvented: a sequence-based pLM can be extended to incorporate the structure modality through continuous tokens, i.e., high-fidelity protein structure latents that avoid vector quantization. Specifically, we propose a hybrid diffusion protein language model, HD-Prot, which embeds a continuous-valued diffusion head atop a discrete pLM, enabling seamless operation with both discrete and continuous tokens for joint sequence-structure modeling. It captures inter-token dependencies across modalities through a unified absorbing diffusion process, and estimates per-token distributions via categorical prediction for sequences and continuous diffusion for structures. Extensive empirical results show that HD-Prot achieves competitive performance in unconditional sequence-structure co-generation, motif-scaffolding, protein structure prediction, and inverse folding tasks, performing on par with state-of-the-art multimodal pLMs despite being developed under limited computational resources. It highlights the viability of simultaneously estimating categorical and continuous distributions within a unified language model architecture, offering a promising alternative direction for multimodal pLMs.
TOMG-Bench: Evaluating LLMs on Text-based Open Molecule Generation
Dec 19, 2024



Abstract:In this paper, we propose Text-based Open Molecule Generation Benchmark (TOMG-Bench), the first benchmark to evaluate the open-domain molecule generation capability of LLMs. TOMG-Bench encompasses a dataset of three major tasks: molecule editing (MolEdit), molecule optimization (MolOpt), and customized molecule generation (MolCustom). Each task further contains three subtasks, with each subtask comprising 5,000 test samples. Given the inherent complexity of open molecule generation, we have also developed an automated evaluation system that helps measure both the quality and the accuracy of the generated molecules. Our comprehensive benchmarking of 25 LLMs reveals the current limitations and potential areas for improvement in text-guided molecule discovery. Furthermore, with the assistance of OpenMolIns, a specialized instruction tuning dataset proposed for solving challenges raised by TOMG-Bench, Llama3.1-8B could outperform all the open-source general LLMs, even surpassing GPT-3.5-turbo by 46.5\% on TOMG-Bench. Our codes and datasets are available through https://github.com/phenixace/TOMG-Bench.
MolReFlect: Towards In-Context Fine-grained Alignments between Molecules and Texts
Nov 22, 2024



Abstract:Molecule discovery is a pivotal research field, impacting everything from the medicines we take to the materials we use. Recently, Large Language Models (LLMs) have been widely adopted in molecule understanding and generation, yet the alignments between molecules and their corresponding captions remain a significant challenge. Previous endeavours often treat the molecule as a general SMILES string or molecular graph, neglecting the fine-grained alignments between the molecular sub-structures and the descriptive textual phrases, which are crucial for accurate and explainable predictions. In this case, we introduce MolReFlect, a novel teacher-student framework designed to contextually perform the molecule-caption alignments in a fine-grained way. Our approach initially leverages a larger teacher LLM to label the detailed alignments by directly extracting critical phrases from molecule captions or SMILES strings and implying them to corresponding sub-structures or characteristics. To refine these alignments, we propose In-Context Selective Reflection, which retrieves previous extraction results as context examples for teacher LLM to reflect and lets a smaller student LLM select from in-context reflection and previous extraction results. Finally, we enhance the learning process of the student LLM through Chain-of-Thought In-Context Molecule Tuning, integrating the fine-grained alignments and the reasoning processes within the Chain-of-Thought format. Our experimental results demonstrate that MolReFlect enables LLMs like Mistral-7B to significantly outperform the previous baselines, achieving SOTA performance on the ChEBI-20 dataset. This advancement not only enhances the generative capabilities of LLMs in the molecule-caption translation task, but also contributes to a more explainable framework.
Recommender Systems in the Era of Large Language Models
Jul 05, 2023Abstract:With the prosperity of e-commerce and web applications, Recommender Systems (RecSys) have become an important component of our daily life, providing personalized suggestions that cater to user preferences. While Deep Neural Networks (DNNs) have made significant advancements in enhancing recommender systems by modeling user-item interactions and incorporating textual side information, DNN-based methods still face limitations, such as difficulties in understanding users' interests and capturing textual side information, inabilities in generalizing to various recommendation scenarios and reasoning on their predictions, etc. Meanwhile, the emergence of Large Language Models (LLMs), such as ChatGPT and GPT4, has revolutionized the fields of Natural Language Processing (NLP) and Artificial Intelligence (AI), due to their remarkable abilities in fundamental responsibilities of language understanding and generation, as well as impressive generalization and reasoning capabilities. As a result, recent studies have attempted to harness the power of LLMs to enhance recommender systems. Given the rapid evolution of this research direction in recommender systems, there is a pressing need for a systematic overview that summarizes existing LLM-empowered recommender systems, to provide researchers in relevant fields with an in-depth understanding. Therefore, in this paper, we conduct a comprehensive review of LLM-empowered recommender systems from various aspects including Pre-training, Fine-tuning, and Prompting. More specifically, we first introduce representative methods to harness the power of LLMs (as a feature encoder) for learning representations of users and items. Then, we review recent techniques of LLMs for enhancing recommender systems from three paradigms, namely pre-training, fine-tuning, and prompting. Finally, we comprehensively discuss future directions in this emerging field.
Empowering Molecule Discovery for Molecule-Caption Translation with Large Language Models: A ChatGPT Perspective
Jun 11, 2023Abstract:Molecule discovery plays a crucial role in various scientific fields, advancing the design of tailored materials and drugs. Traditional methods for molecule discovery follow a trial-and-error process, which are both time-consuming and costly, while computational approaches such as artificial intelligence (AI) have emerged as revolutionary tools to expedite various tasks, like molecule-caption translation. Despite the importance of molecule-caption translation for molecule discovery, most of the existing methods heavily rely on domain experts, require excessive computational cost, and suffer from poor performance. On the other hand, Large Language Models (LLMs), like ChatGPT, have shown remarkable performance in various cross-modal tasks due to their great powerful capabilities in natural language understanding, generalization, and reasoning, which provides unprecedented opportunities to advance molecule discovery. To address the above limitations, in this work, we propose a novel LLMs-based framework (\textbf{MolReGPT}) for molecule-caption translation, where a retrieval-based prompt paradigm is introduced to empower molecule discovery with LLMs like ChatGPT without fine-tuning. More specifically, MolReGPT leverages the principle of molecular similarity to retrieve similar molecules and their text descriptions from a local database to ground the generation of LLMs through in-context few-shot molecule learning. We evaluate the effectiveness of MolReGPT via molecule-caption translation, which includes molecule understanding and text-based molecule generation. Experimental results show that MolReGPT outperforms fine-tuned models like MolT5-base without any additional training. To the best of our knowledge, MolReGPT is the first work to leverage LLMs in molecule-caption translation for advancing molecule discovery.
Improving User Controlled Table-To-Text Generation Robustness
Feb 20, 2023



Abstract:In this work we study user controlled table-to-text generation where users explore the content in a table by selecting cells and reading a natural language description thereof automatically produce by a natural language generator. Such generation models usually learn from carefully selected cell combinations (clean cell selections); however, in practice users may select unexpected, redundant, or incoherent cell combinations (noisy cell selections). In experiments, we find that models perform well on test sets coming from the same distribution as the train data but their performance drops when evaluated on realistic noisy user inputs. We propose a fine-tuning regime with additional user-simulated noisy cell selections. Models fine-tuned with the proposed regime gain 4.85 BLEU points on user noisy test cases and 1.4 on clean test cases; and achieve comparable state-of-the-art performance on the ToTTo dataset.
Generative Diffusion Models on Graphs: Methods and Applications
Feb 06, 2023Abstract:Diffusion models, as a novel generative paradigm, have achieved remarkable success in various image generation tasks such as image inpainting, image-to-text translation, and video generation. Graph generation is a crucial computational task on graphs with numerous real-world applications. It aims to learn the distribution of given graphs and then generate new graphs. Given the great success of diffusion models in image generation, increasing efforts have been made to leverage these techniques to advance graph generation in recent years. In this paper, we first provide a comprehensive overview of generative diffusion models on graphs, In particular, we review representative algorithms for three variants of graph diffusion models, i.e., Score Matching with Langevin Dynamics (SMLD), Denoising Diffusion Probabilistic Model (DDPM), and Score-based Generative Model (SGM). Then, we summarize the major applications of generative diffusion models on graphs with a specific focus on molecule and protein modeling. Finally, we discuss promising directions in generative diffusion models on graph-structured data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge