Yujie Qiao
P-FOLIO: Evaluating and Improving Logical Reasoning with Abundant Human-Written Reasoning Chains
Oct 11, 2024



Abstract:Existing methods on understanding the capabilities of LLMs in logical reasoning rely on binary entailment classification or synthetically derived rationales, which are not sufficient for proper investigation of model's capabilities. We present P-FOLIO, a human-annotated dataset consisting of diverse and complex reasoning chains for a set of realistic logical reasoning stories also written by humans. P-FOLIO is collected with an annotation protocol that facilitates humans to annotate well-structured natural language proofs for first-order logic reasoning problems in a step-by-step manner. The number of reasoning steps in P-FOLIO span from 0 to 20. We further use P-FOLIO to evaluate and improve large-language-model (LLM) reasoning capabilities. We evaluate LLM reasoning capabilities at a fine granularity via single-step inference rule classification, with more diverse inference rules of more diverse and higher levels of complexities than previous works. Given that a single model-generated reasoning chain could take a completely different path than the human-annotated one, we sample multiple reasoning chains from a model and use pass@k metrics for evaluating the quality of model-generated reasoning chains. We show that human-written reasoning chains significantly boost the logical reasoning capabilities of LLMs via many-shot prompting and fine-tuning. Furthermore, fine-tuning Llama3-7B on P-FOLIO improves the model performance by 10% or more on three other out-of-domain logical reasoning datasets. We also conduct detailed analysis to show where most powerful LLMs fall short in reasoning. We will release the dataset and code publicly.
MedGen: A Python Natural Language Processing Toolkit for Medical Text Processing
Nov 28, 2023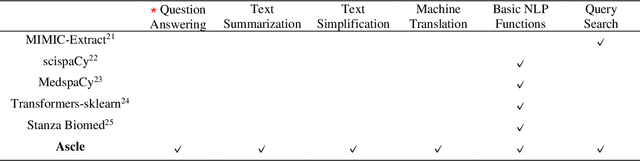
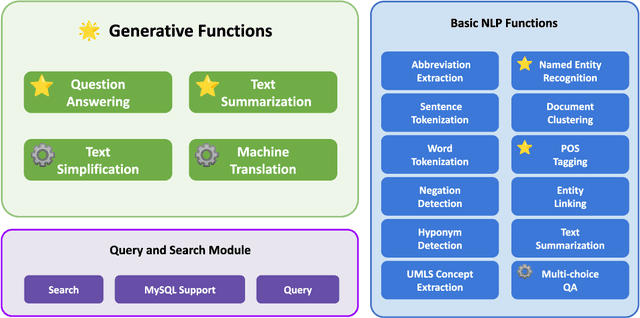
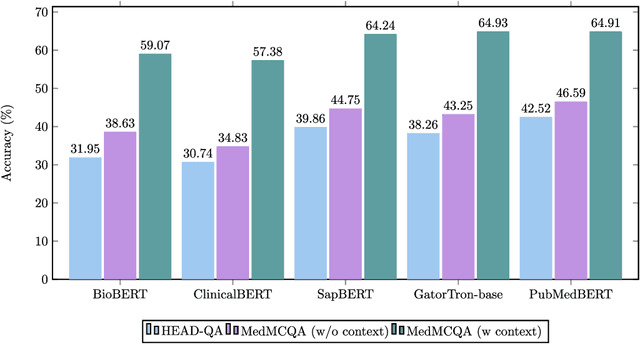
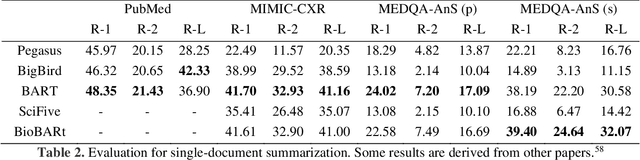
Abstract:This study introduces MedGen, a comprehensive natural language processing (NLP) toolkit designed for medical text processing. MedGen is tailored for biomedical researchers and healthcare professionals with an easy-to-use, all-in-one solution that requires minimal programming expertise. It includes (1) Generative Functions: For the first time, MedGen includes four advanced generative functions: question answering, text summarization, text simplification, and machine translation; (2) Basic NLP Functions: MedGen integrates 12 essential NLP functions such as word tokenization and sentence segmentation; and (3) Query and Search Capabilities: MedGen provides user-friendly query and search functions on text corpora. We fine-tuned 32 domain-specific language models, evaluated them thoroughly on 24 established benchmarks and conducted manual reviews with clinicians. Additionally, we expanded our toolkit by introducing query and search functions, while also standardizing and integrating functions from third-party libraries. The toolkit, its models, and associated data are publicly available via https://github.com/Yale-LILY/MedGen.
FOLIO: Natural Language Reasoning with First-Order Logic
Sep 02, 2022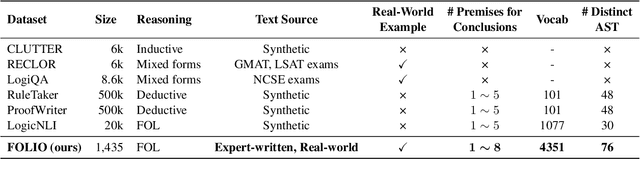
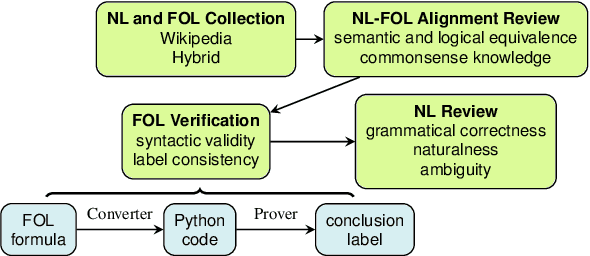
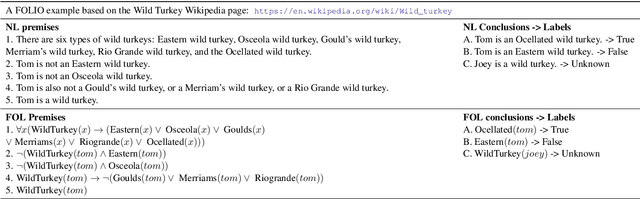
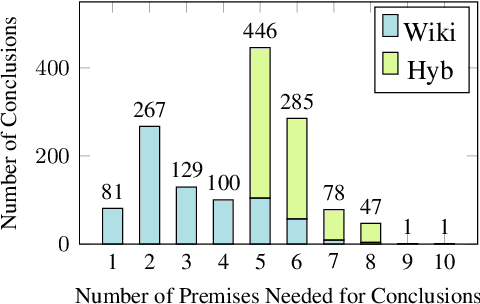
Abstract:We present FOLIO, a human-annotated, open-domain, and logically complex and diverse dataset for reasoning in natural language (NL), equipped with first order logic (FOL) annotations. FOLIO consists of 1,435 examples (unique conclusions), each paired with one of 487 sets of premises which serve as rules to be used to deductively reason for the validity of each conclusion. The logical correctness of premises and conclusions is ensured by their parallel FOL annotations, which are automatically verified by our FOL inference engine. In addition to the main NL reasoning task, NL-FOL pairs in FOLIO automatically constitute a new NL-FOL translation dataset using FOL as the logical form. Our experiments on FOLIO systematically evaluate the FOL reasoning ability of supervised fine-tuning on medium-sized language models (BERT, RoBERTa) and few-shot prompting on large language models (GPT-NeoX, OPT, GPT-3, Codex). For NL-FOL translation, we experiment with GPT-3 and Codex. Our results show that one of the most capable Large Language Model (LLM) publicly available, GPT-3 davinci, achieves only slightly better than random results with few-shot prompting on a subset of FOLIO, and the model is especially bad at predicting the correct truth values for False and Unknown conclusions. Our dataset and code are available at https://github.com/Yale-LILY/FOLIO.
EHRKit: A Python Natural Language Processing Toolkit for Electronic Health Record Texts
Apr 13, 2022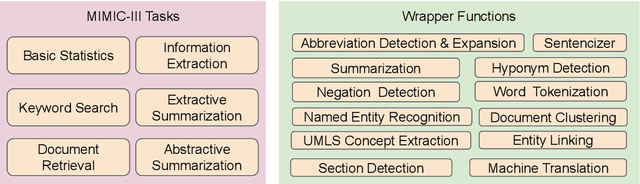
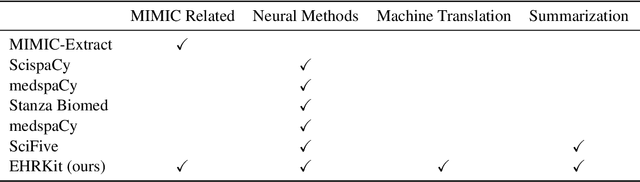

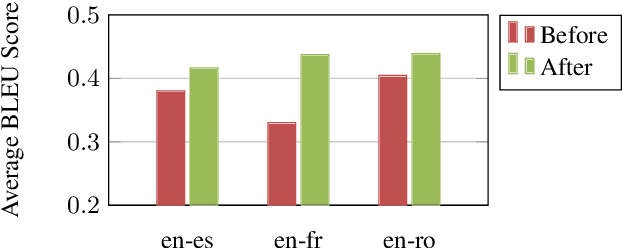
Abstract:The Electronic Health Record (EHR) is an essential part of the modern medical system and impacts healthcare delivery, operations, and research. Unstructured text is attracting much attention despite structured information in the EHRs and has become an exciting research field. The success of the recent neural Natural Language Processing (NLP) method has led to a new direction for processing unstructured clinical notes. In this work, we create a python library for clinical texts, EHRKit. This library contains two main parts: MIMIC-III-specific functions and tasks specific functions. The first part introduces a list of interfaces for accessing MIMIC-III NOTEEVENTS data, including basic search, information retrieval, and information extraction. The second part integrates many third-party libraries for up to 12 off-shelf NLP tasks such as named entity recognition, summarization, machine translation, etc.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge