Yi Hong
Harmonizing Generalization and Specialization: Uncertainty-Informed Collaborative Learning for Semi-supervised Medical Image Segmentation
Dec 15, 2025Abstract:Vision foundation models have demonstrated strong generalization in medical image segmentation by leveraging large-scale, heterogeneous pretraining. However, they often struggle to generalize to specialized clinical tasks under limited annotations or rare pathological variations, due to a mismatch between general priors and task-specific requirements. To address this, we propose Uncertainty-informed Collaborative Learning (UnCoL), a dual-teacher framework that harmonizes generalization and specialization in semi-supervised medical image segmentation. Specifically, UnCoL distills both visual and semantic representations from a frozen foundation model to transfer general knowledge, while concurrently maintaining a progressively adapting teacher to capture fine-grained and task-specific representations. To balance guidance from both teachers, pseudo-label learning in UnCoL is adaptively regulated by predictive uncertainty, which selectively suppresses unreliable supervision and stabilizes learning in ambiguous regions. Experiments on diverse 2D and 3D segmentation benchmarks show that UnCoL consistently outperforms state-of-the-art semi-supervised methods and foundation model baselines. Moreover, our model delivers near fully supervised performance with markedly reduced annotation requirements.
Enhancing 3D Medical Image Understanding with Pretraining Aided by 2D Multimodal Large Language Models
Sep 11, 2025Abstract:Understanding 3D medical image volumes is critical in the medical field, yet existing 3D medical convolution and transformer-based self-supervised learning (SSL) methods often lack deep semantic comprehension. Recent advancements in multimodal large language models (MLLMs) provide a promising approach to enhance image understanding through text descriptions. To leverage these 2D MLLMs for improved 3D medical image understanding, we propose Med3DInsight, a novel pretraining framework that integrates 3D image encoders with 2D MLLMs via a specially designed plane-slice-aware transformer module. Additionally, our model employs a partial optimal transport based alignment, demonstrating greater tolerance to noise introduced by potential noises in LLM-generated content. Med3DInsight introduces a new paradigm for scalable multimodal 3D medical representation learning without requiring human annotations. Extensive experiments demonstrate our state-of-the-art performance on two downstream tasks, i.e., segmentation and classification, across various public datasets with CT and MRI modalities, outperforming current SSL methods. Med3DInsight can be seamlessly integrated into existing 3D medical image understanding networks, potentially enhancing their performance. Our source code, generated datasets, and pre-trained models will be available at https://github.com/Qybc/Med3DInsight.
HoloDx: Knowledge- and Data-Driven Multimodal Diagnosis of Alzheimer's Disease
Apr 27, 2025



Abstract:Accurate diagnosis of Alzheimer's disease (AD) requires effectively integrating multimodal data and clinical expertise. However, existing methods often struggle to fully utilize multimodal information and lack structured mechanisms to incorporate dynamic domain knowledge. To address these limitations, we propose HoloDx, a knowledge- and data-driven framework that enhances AD diagnosis by aligning domain knowledge with multimodal clinical data. HoloDx incorporates a knowledge injection module with a knowledge-aware gated cross-attention, allowing the model to dynamically integrate domain-specific insights from both large language models (LLMs) and clinical expertise. Also, a memory injection module with a designed prototypical memory attention enables the model to retain and retrieve subject-specific information, ensuring consistency in decision-making. By jointly leveraging these mechanisms, HoloDx enhances interpretability, improves robustness, and effectively aligns prior knowledge with current subject data. Evaluations on five AD datasets demonstrate that HoloDx outperforms state-of-the-art methods, achieving superior diagnostic accuracy and strong generalization across diverse cohorts. The source code will be released upon publication acceptance.
An Event-centric Framework for Predicting Crime Hotspots with Flexible Time Intervals
Nov 02, 2024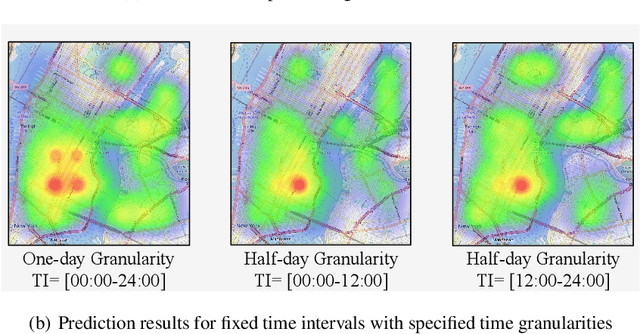
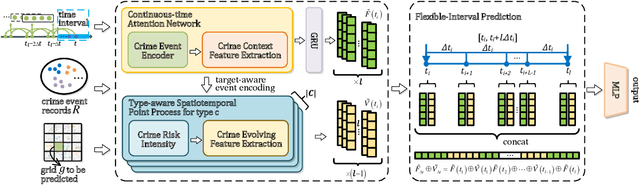

Abstract:Predicting crime hotspots in a city is a complex and critical task with significant societal implications. Numerous spatiotemporal correlations and irregularities pose substantial challenges to this endeavor. Existing methods commonly employ fixed-time granularities and sequence prediction models. However, determining appropriate time granularities is difficult, leading to inaccurate predictions for specific time windows. For example, users might ask: What are the crime hotspots during 12:00-20:00? To address this issue, we introduce FlexiCrime, a novel event-centric framework for predicting crime hotspots with flexible time intervals. FlexiCrime incorporates a continuous-time attention network to capture correlations between crime events, which learns crime context features, representing general crime patterns across time points and locations. Furthermore, we introduce a type-aware spatiotemporal point process that learns crime-evolving features, measuring the risk of specific crime types at a given time and location by considering the frequency of past crime events. The crime context and evolving features together allow us to predict whether an urban area is a crime hotspot given a future time interval. To evaluate FlexiCrime's effectiveness, we conducted experiments using real-world datasets from two cities, covering twelve crime types. The results show that our model outperforms baseline techniques in predicting crime hotspots over flexible time intervals.
NODER: Image Sequence Regression Based on Neural Ordinary Differential Equations
Jul 18, 2024



Abstract:Regression on medical image sequences can capture temporal image pattern changes and predict images at missing or future time points. However, existing geodesic regression methods limit their regression performance by a strong underlying assumption of linear dynamics, while diffusion-based methods have high computational costs and lack constraints to preserve image topology. In this paper, we propose an optimization-based new framework called NODER, which leverages neural ordinary differential equations to capture complex underlying dynamics and reduces its high computational cost of handling high-dimensional image volumes by introducing the latent space. We compare our NODER with two recent regression methods, and the experimental results on ADNI and ACDC datasets demonstrate that our method achieves the state-of-the-art performance in 3D image regression. Our model needs only a couple of images in a sequence for prediction, which is practical, especially for clinical situations where extremely limited image time series are available for analysis. Our source code is available at https://github.com/ZedKing12138/NODER-pytorch.
OTFS Channel Estimation and Detection for Channels with Very Large Delay Spread
Apr 12, 2024Abstract:In low latency applications and in general, for overspread channels, channel delay spread is a large percentage of the transmission frame duration. In this paper, we consider OTFS in an overspread channel exhibiting a delay spread that exceeds the block duration in a frame, where traditional channel estimation (CE) fails. We propose a two-stage CE method based on a delay-Doppler (DD) training frame, consisting of a dual chirp converted from time domain and a higher power pilot. The first stage employs a DD domain embedded pilot CE to estimate the aliased delays (due to modulo operation) and Doppler shifts, followed by identifying all the underspread paths not coinciding with any overspread path. The second stage utilizes time domain dual chirp correlation to estimate the actual delays and Doppler shifts of the remaining paths. This stage also resolves ambiguity in estimating delays and Doppler shifts for paths sharing same aliased delay. Furthermore, we present a modified low-complexity maximum ratio combining (MRC) detection algorithm for OTFS in overspread channels. Finally, we evaluate performance of OTFS using the proposed CE and the modified MRC detection in terms of normalized mean square error (NMSE) and bit error rate (BER).
Med3DInsight: Enhancing 3D Medical Image Understanding with 2D Multi-Modal Large Language Models
Mar 08, 2024


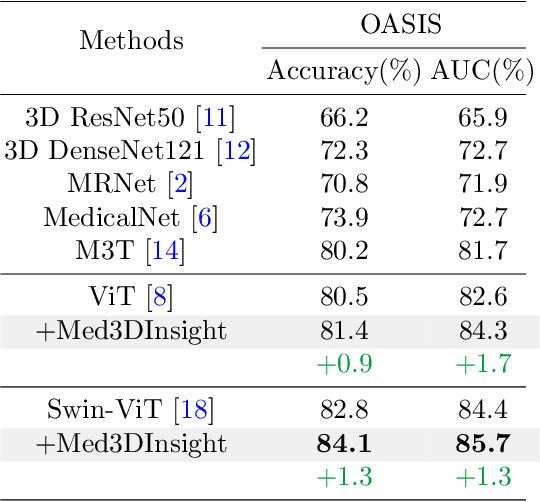
Abstract:Understanding 3D medical image volumes is a critical task in the medical domain. However, existing 3D convolution and transformer-based methods have limited semantic understanding of an image volume and also need a large set of volumes for training. Recent advances in multi-modal large language models (MLLMs) provide a new and promising way to understand images with the help of text descriptions. However, most current MLLMs are designed for 2D natural images. To enhance the 3D medical image understanding with 2D MLLMs, we propose a novel pre-training framework called Med3DInsight, which marries existing 3D image encoders with 2D MLLMs and bridges them via a designed Plane-Slice-Aware Transformer (PSAT) module. Extensive experiments demonstrate our SOTA performance on two downstream segmentation and classification tasks, including three public datasets with CT and MRI modalities and comparison to more than ten baselines. Med3DInsight can be easily integrated into any current 3D medical image understanding network and improves its performance by a good margin.
AliFuse: Aligning and Fusing Multi-modal Medical Data for Computer-Aided Diagnosis
Jan 07, 2024



Abstract:Medical data collected for making a diagnostic decision are typically multi-modal and provide complementary perspectives of a subject. A computer-aided diagnosis system welcomes multi-modal inputs; however, how to effectively fuse such multi-modal data is a challenging task and attracts a lot of attention in the medical research field. In this paper, we propose a transformer-based framework, called Alifuse, for aligning and fusing multi-modal medical data. Specifically, we convert images and unstructured and structured texts into vision and language tokens, and use intramodal and intermodal attention mechanisms to learn holistic representations of all imaging and non-imaging data for classification. We apply Alifuse to classify Alzheimer's disease and obtain state-of-the-art performance on five public datasets, by outperforming eight baselines. The source code will be available online later.
Volumetric Medical Image Segmentation via Scribble Annotations and Shape Priors
Oct 12, 2023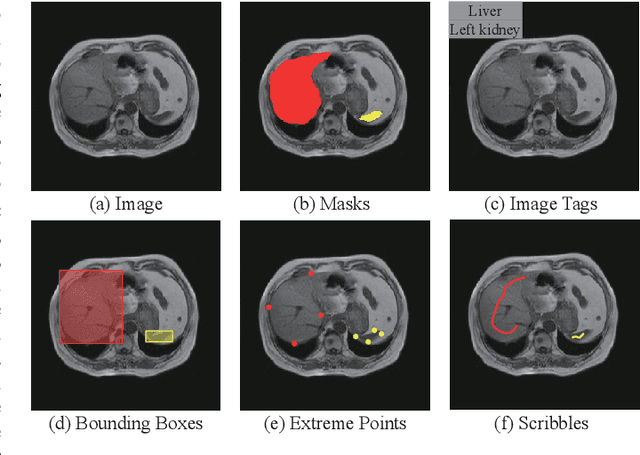
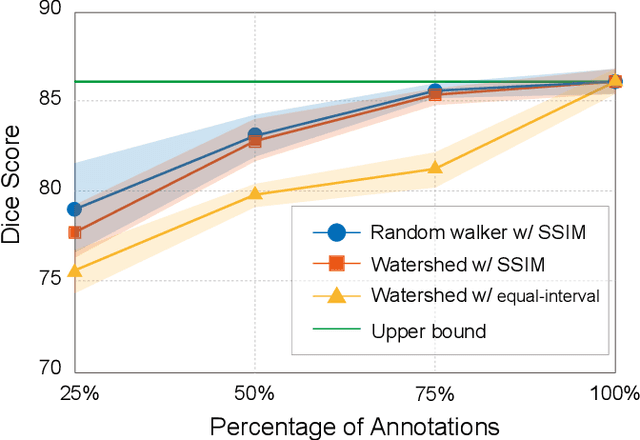
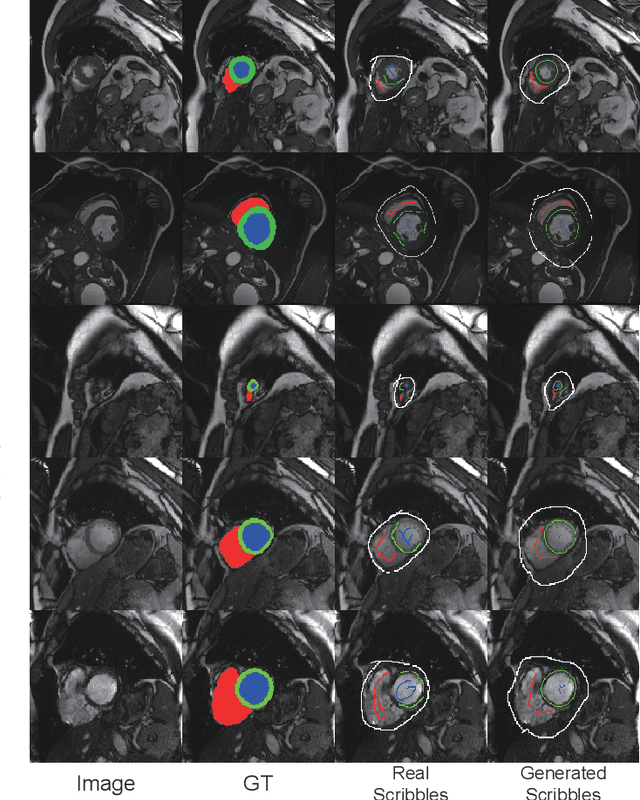
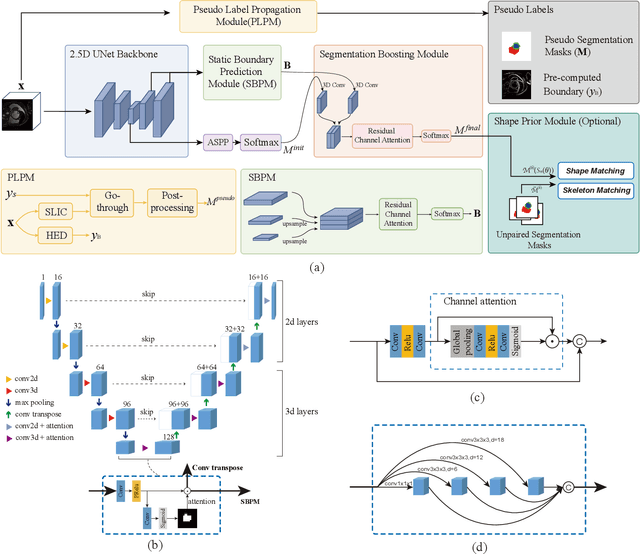
Abstract:Recently, weakly-supervised image segmentation using weak annotations like scribbles has gained great attention in computer vision and medical image analysis, since such annotations are much easier to obtain compared to time-consuming and labor-intensive labeling at the pixel/voxel level. However, due to a lack of structure supervision on regions of interest (ROIs), existing scribble-based methods suffer from poor boundary localization. Furthermore, most current methods are designed for 2D image segmentation, which do not fully leverage the volumetric information if directly applied to each image slice. In this paper, we propose a scribble-based volumetric image segmentation, Scribble2D5, which tackles 3D anisotropic image segmentation and aims to its improve boundary prediction. To achieve this, we augment a 2.5D attention UNet with a proposed label propagation module to extend semantic information from scribbles and use a combination of static and active boundary prediction to learn ROI's boundary and regularize its shape. Also, we propose an optional add-on component, which incorporates the shape prior information from unpaired segmentation masks to further improve model accuracy. Extensive experiments on three public datasets and one private dataset demonstrate our Scribble2D5 achieves state-of-the-art performance on volumetric image segmentation using scribbles and shape prior if available.
MedBLIP: Bootstrapping Language-Image Pre-training from 3D Medical Images and Texts
May 18, 2023



Abstract:Vision-language pre-training (VLP) models have been demonstrated to be effective in many computer vision applications. In this paper, we consider developing a VLP model in the medical domain for making computer-aided diagnoses (CAD) based on image scans and text descriptions in electronic health records, as done in practice. To achieve our goal, we present a lightweight CAD system MedBLIP, a new paradigm for bootstrapping VLP from off-the-shelf frozen pre-trained image encoders and frozen large language models. We design a MedQFormer module to bridge the gap between 3D medical images and 2D pre-trained image encoders and language models as well. To evaluate the effectiveness of our MedBLIP, we collect more than 30,000 image volumes from five public Alzheimer's disease (AD) datasets, i.e., ADNI, NACC, OASIS, AIBL, and MIRIAD. On this largest AD dataset we know, our model achieves the SOTA performance on the zero-shot classification of healthy, mild cognitive impairment (MCI), and AD subjects, and shows its capability of making medical visual question answering (VQA). The code and pre-trained models is available online: https://github.com/Qybc/MedBLIP.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge