Xin Yue
Towards Video to Piano Music Generation with Chain-of-Perform Support Benchmarks
May 26, 2025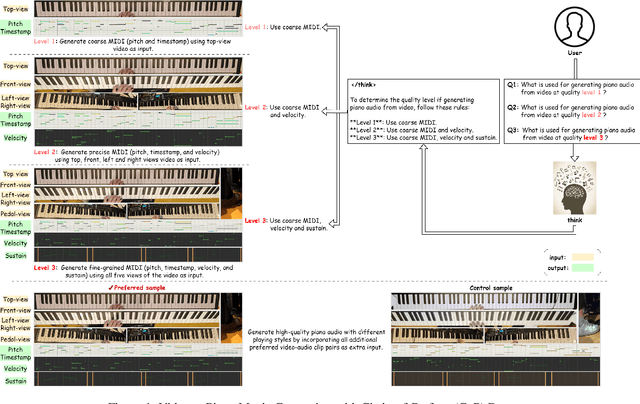
Abstract:Generating high-quality piano audio from video requires precise synchronization between visual cues and musical output, ensuring accurate semantic and temporal alignment.However, existing evaluation datasets do not fully capture the intricate synchronization required for piano music generation. A comprehensive benchmark is essential for two primary reasons: (1) existing metrics fail to reflect the complexity of video-to-piano music interactions, and (2) a dedicated benchmark dataset can provide valuable insights to accelerate progress in high-quality piano music generation. To address these challenges, we introduce the CoP Benchmark Dataset-a fully open-sourced, multimodal benchmark designed specifically for video-guided piano music generation. The proposed Chain-of-Perform (CoP) benchmark offers several compelling features: (1) detailed multimodal annotations, enabling precise semantic and temporal alignment between video content and piano audio via step-by-step Chain-of-Perform guidance; (2) a versatile evaluation framework for rigorous assessment of both general-purpose and specialized video-to-piano generation tasks; and (3) full open-sourcing of the dataset, annotations, and evaluation protocols. The dataset is publicly available at https://github.com/acappemin/Video-to-Audio-and-Piano, with a continuously updated leaderboard to promote ongoing research in this domain.
HemSeg-200: A Voxel-Annotated Dataset for Intracerebral Hemorrhages Segmentation in Brain CT Scans
May 23, 2024Abstract:Acute intracerebral hemorrhage is a life-threatening condition that demands immediate medical intervention. Intraparenchymal hemorrhage (IPH) and intraventricular hemorrhage (IVH) are critical subtypes of this condition. Clinically, when such hemorrhages are suspected, immediate CT scanning is essential to assess the extent of the bleeding and to facilitate the formulation of a targeted treatment plan. While current research in deep learning has largely focused on qualitative analyses, such as identifying subtypes of cerebral hemorrhages, there remains a significant gap in quantitative analysis crucial for enhancing clinical treatments. Addressing this gap, our paper introduces a dataset comprising 222 CT annotations, sourced from the RSNA 2019 Brain CT Hemorrhage Challenge and meticulously annotated at the voxel level for precise IPH and IVH segmentation. This dataset was utilized to train and evaluate seven advanced medical image segmentation algorithms, with the goal of refining the accuracy of segmentation for these hemorrhages. Our findings demonstrate that this dataset not only furthers the development of sophisticated segmentation algorithms but also substantially aids scientific research and clinical practice by improving the diagnosis and management of these severe hemorrhages. Our dataset and codes are available at \url{https://github.com/songchangwei/3DCT-SD-IVH-ICH}.
Comparative Analysis of ImageNet Pre-Trained Deep Learning Models and DINOv2 in Medical Imaging Classification
Feb 13, 2024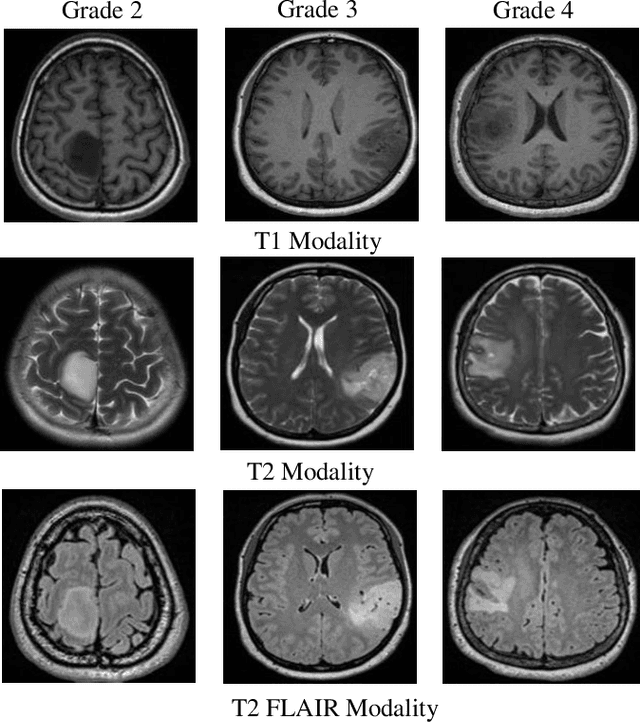
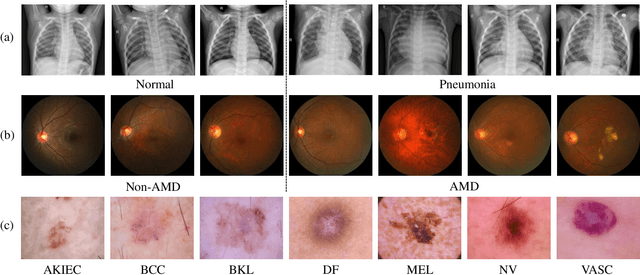
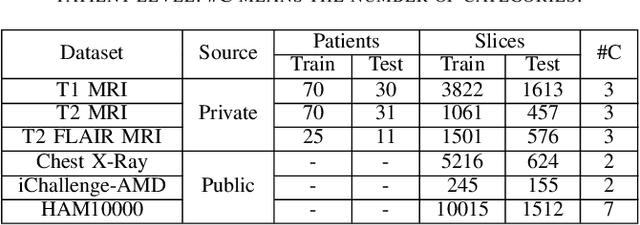
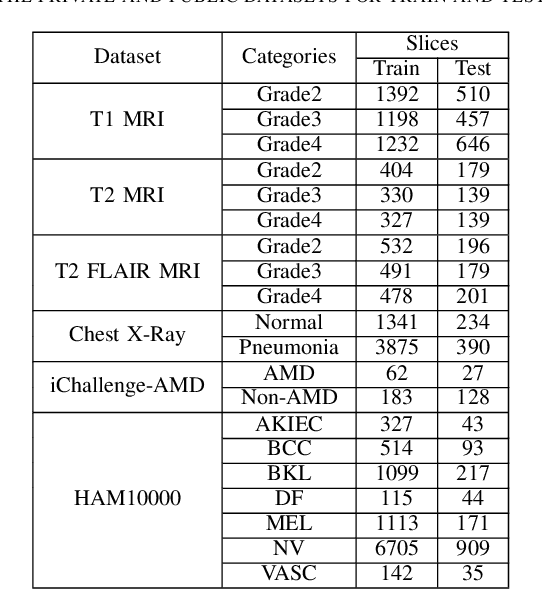
Abstract:Medical image analysis frequently encounters data scarcity challenges. Transfer learning has been effective in addressing this issue while conserving computational resources. The recent advent of foundational models like the DINOv2, which uses the vision transformer architecture, has opened new opportunities in the field and gathered significant interest. However, DINOv2's performance on clinical data still needs to be verified. In this paper, we performed a glioma grading task using three clinical modalities of brain MRI data. We compared the performance of various pre-trained deep learning models, including those based on ImageNet and DINOv2, in a transfer learning context. Our focus was on understanding the impact of the freezing mechanism on performance. We also validated our findings on three other types of public datasets: chest radiography, fundus radiography, and dermoscopy. Our findings indicate that in our clinical dataset, DINOv2's performance was not as strong as ImageNet-based pre-trained models, whereas in public datasets, DINOv2 generally outperformed other models, especially when using the frozen mechanism. Similar performance was observed with various sizes of DINOv2 models across different tasks. In summary, DINOv2 is viable for medical image classification tasks, particularly with data resembling natural images. However, its effectiveness may vary with data that significantly differs from natural images such as MRI. In addition, employing smaller versions of the model can be adequate for medical task, offering resource-saving benefits. Our codes are available at https://github.com/GuanghuiFU/medical_DINOv2_eval.
Morphology-Enhanced CAM-Guided SAM for weakly supervised Breast Lesion Segmentation
Nov 18, 2023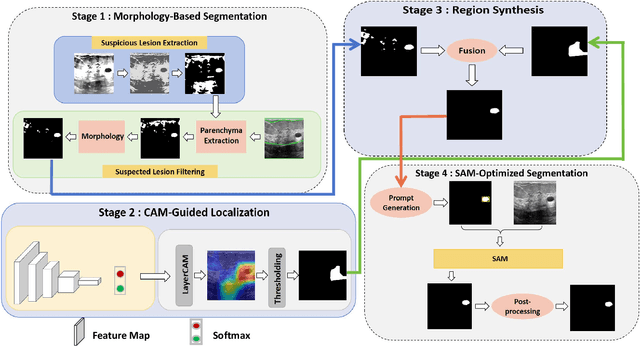
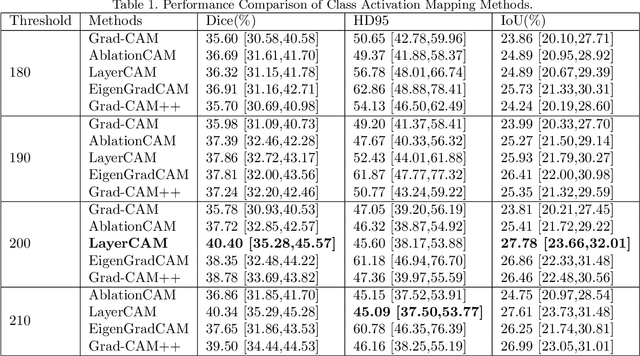
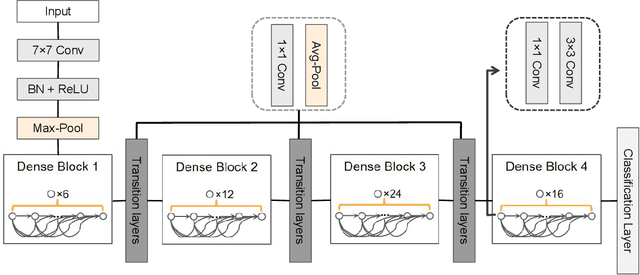

Abstract:Breast cancer diagnosis challenges both patients and clinicians, with early detection being crucial for effective treatment. Ultrasound imaging plays a key role in this, but its utility is hampered by the need for precise lesion segmentation-a task that is both time-consuming and labor-intensive. To address these challenges, we propose a new framework: a morphology-enhanced, Class Activation Map (CAM)-guided model, which is optimized using a computer vision foundation model known as SAM. This innovative framework is specifically designed for weakly supervised lesion segmentation in early-stage breast ultrasound images. Our approach uniquely leverages image-level annotations, which removes the requirement for detailed pixel-level annotation. Initially, we perform a preliminary segmentation using breast lesion morphology knowledge. Following this, we accurately localize lesions by extracting semantic information through a CAM-based heatmap. These two elements are then fused together, serving as a prompt to guide the SAM in performing refined segmentation. Subsequently, post-processing techniques are employed to rectify topological errors made by the SAM. Our method not only simplifies the segmentation process but also attains accuracy comparable to supervised learning methods that rely on pixel-level annotation. Our framework achieves a Dice score of 74.39% on the test set, demonstrating compareable performance with supervised learning methods. Additionally, it outperforms a supervised learning model, in terms of the Hausdorff distance, scoring 24.27 compared to Deeplabv3+'s 32.22. These experimental results showcase its feasibility and superior performance in integrating weakly supervised learning with SAM. The code is made available at: https://github.com/YueXin18/MorSeg-CAM-SAM.
Ultrafast CMOS image sensors and data-enabled super-resolution for multimodal radiographic imaging and tomography
Jan 27, 2023
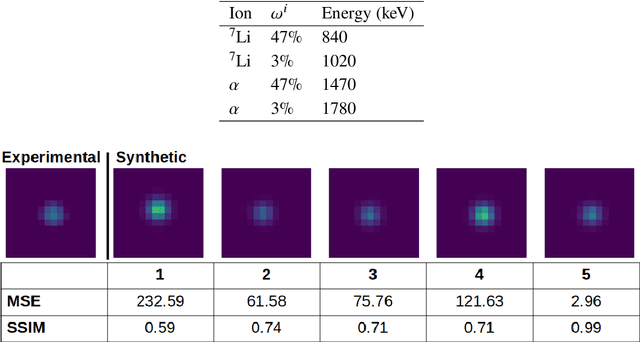
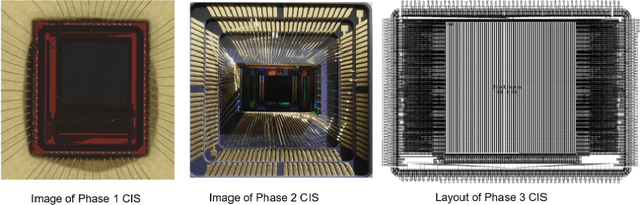
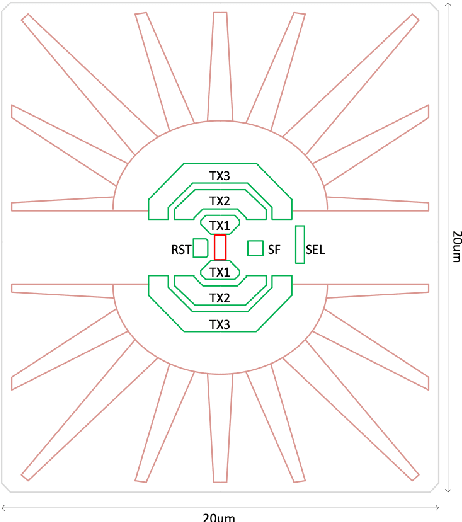
Abstract:We summarize recent progress in ultrafast Complementary Metal Oxide Semiconductor (CMOS) image sensor development and the application of neural networks for post-processing of CMOS and charge-coupled device (CCD) image data to achieve sub-pixel resolution (thus $super$-$resolution$). The combination of novel CMOS pixel designs and data-enabled image post-processing provides a promising path towards ultrafast high-resolution multi-modal radiographic imaging and tomography applications.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge