Xiaodi Huang
Multi-label feature selection based on binary hashing learning and dynamic graph constraints
Mar 18, 2025



Abstract:Multi-label learning poses significant challenges in extracting reliable supervisory signals from the label space. Existing approaches often employ continuous pseudo-labels to replace binary labels, improving supervisory information representation. However, these methods can introduce noise from irrelevant labels and lead to unreliable graph structures. To overcome these limitations, this study introduces a novel multi-label feature selection method called Binary Hashing and Dynamic Graph Constraint (BHDG), the first method to integrate binary hashing into multi-label learning. BHDG utilizes low-dimensional binary hashing codes as pseudo-labels to reduce noise and improve representation robustness. A dynamically constrained sample projection space is constructed based on the graph structure of these binary pseudo-labels, enhancing the reliability of the dynamic graph. To further enhance pseudo-label quality, BHDG incorporates label graph constraints and inner product minimization within the sample space. Additionally, an $l_{2,1}$-norm regularization term is added to the objective function to facilitate the feature selection process. The augmented Lagrangian multiplier (ALM) method is employed to optimize binary variables effectively. Comprehensive experiments on 10 benchmark datasets demonstrate that BHDG outperforms ten state-of-the-art methods across six evaluation metrics. BHDG achieves the highest overall performance ranking, surpassing the next-best method by an average of at least 2.7 ranks per metric, underscoring its effectiveness and robustness in multi-label feature selection.
VANER: Leveraging Large Language Model for Versatile and Adaptive Biomedical Named Entity Recognition
Apr 27, 2024Abstract:Prevalent solution for BioNER involves using representation learning techniques coupled with sequence labeling. However, such methods are inherently task-specific, demonstrate poor generalizability, and often require dedicated model for each dataset. To leverage the versatile capabilities of recently remarkable large language models (LLMs), several endeavors have explored generative approaches to entity extraction. Yet, these approaches often fall short of the effectiveness of previouly sequence labeling approaches. In this paper, we utilize the open-sourced LLM LLaMA2 as the backbone model, and design specific instructions to distinguish between different types of entities and datasets. By combining the LLM's understanding of instructions with sequence labeling techniques, we use mix of datasets to train a model capable of extracting various types of entities. Given that the backbone LLMs lacks specialized medical knowledge, we also integrate external entity knowledge bases and employ instruction tuning to compel the model to densely recognize carefully curated entities. Our model VANER, trained with a small partition of parameters, significantly outperforms previous LLMs-based models and, for the first time, as a model based on LLM, surpasses the majority of conventional state-of-the-art BioNER systems, achieving the highest F1 scores across three datasets.
ChatPRCS: A Personalized Support System for English Reading Comprehension based on ChatGPT
Sep 25, 2023



Abstract:As a common approach to learning English, reading comprehension primarily entails reading articles and answering related questions. However, the complexity of designing effective exercises results in students encountering standardized questions, making it challenging to align with individualized learners' reading comprehension ability. By leveraging the advanced capabilities offered by large language models, exemplified by ChatGPT, this paper presents a novel personalized support system for reading comprehension, referred to as ChatPRCS, based on the Zone of Proximal Development theory. ChatPRCS employs methods including reading comprehension proficiency prediction, question generation, and automatic evaluation, among others, to enhance reading comprehension instruction. First, we develop a new algorithm that can predict learners' reading comprehension abilities using their historical data as the foundation for generating questions at an appropriate level of difficulty. Second, a series of new ChatGPT prompt patterns is proposed to address two key aspects of reading comprehension objectives: question generation, and automated evaluation. These patterns further improve the quality of generated questions. Finally, by integrating personalized ability and reading comprehension prompt patterns, ChatPRCS is systematically validated through experiments. Empirical results demonstrate that it provides learners with high-quality reading comprehension questions that are broadly aligned with expert-crafted questions at a statistical level.
GoSum: Extractive Summarization of Long Documents by Reinforcement Learning and Graph Organized discourse state
Nov 18, 2022



Abstract:Handling long texts with structural information and excluding redundancy between summary sentences are essential in extractive document summarization. In this work, we propose GoSum, a novel reinforcement-learning-based extractive model for long-paper summarization. GoSum encodes states by building a heterogeneous graph from different discourse levels for each input document. We evaluate the model on two datasets of scientific articles summarization: PubMed and arXiv where it outperforms all extractive summarization models and most of the strong abstractive baselines.
Mixed Supervision Learning for Whole Slide Image Classification
Jul 05, 2021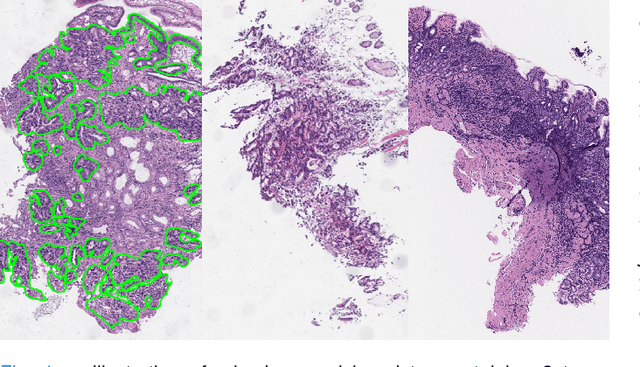
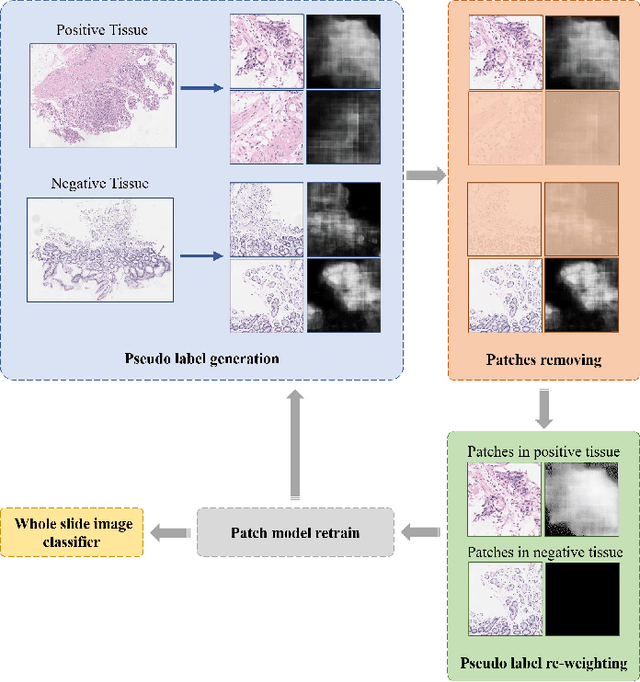
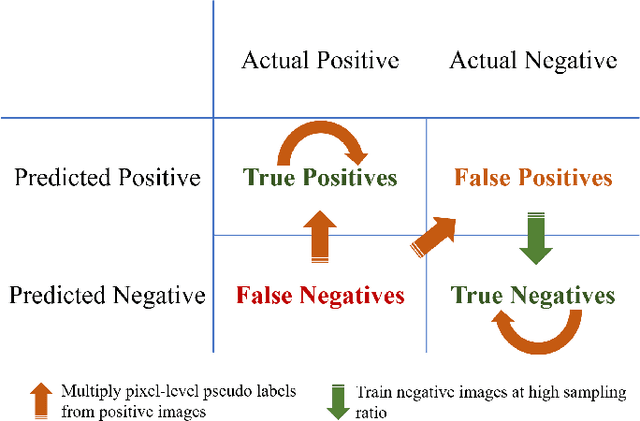
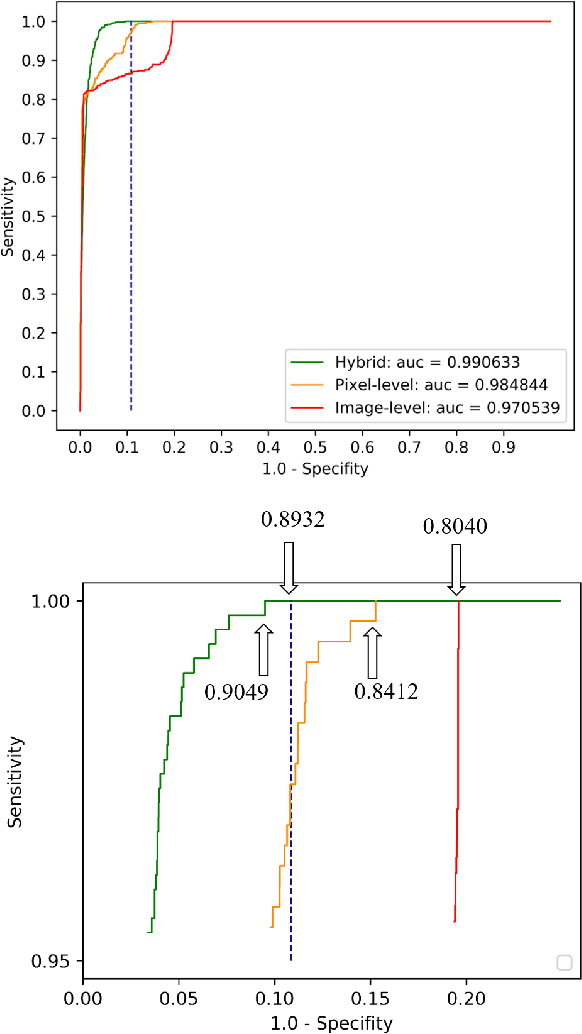
Abstract:Weak supervision learning on classification labels has demonstrated high performance in various tasks. When a few pixel-level fine annotations are also affordable, it is natural to leverage both of the pixel-level (e.g., segmentation) and image level (e.g., classification) annotation to further improve the performance. In computational pathology, however, such weak or mixed supervision learning is still a challenging task, since the high resolution of whole slide images makes it unattainable to perform end-to-end training of classification models. An alternative approach is to analyze such data by patch-base model training, i.e., using self-supervised learning to generate pixel-level pseudo labels for patches. However, such methods usually have model drifting issues, i.e., hard to converge, because the noise accumulates during the self-training process. To handle those problems, we propose a mixed supervision learning framework for super high-resolution images to effectively utilize their various labels (e.g., sufficient image-level coarse annotations and a few pixel-level fine labels). During the patch training stage, this framework can make use of coarse image-level labels to refine self-supervised learning and generate high-quality pixel-level pseudo labels. A comprehensive strategy is proposed to suppress pixel-level false positives and false negatives. Three real-world datasets with very large number of images (i.e., more than 10,000 whole slide images) and various types of labels are used to evaluate the effectiveness of mixed supervision learning. We reduced the false positive rate by around one third compared to state of the art while retaining 100% sensitivity, in the task of image-level classification.
Large-scale Gastric Cancer Screening and Localization Using Multi-task Deep Neural Network
Oct 12, 2019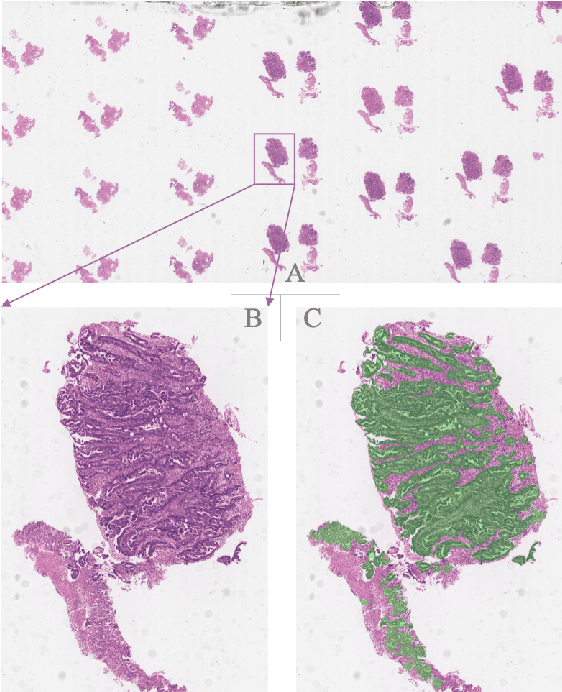
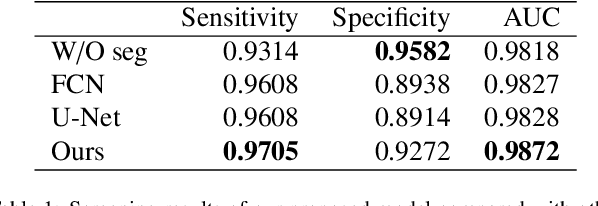
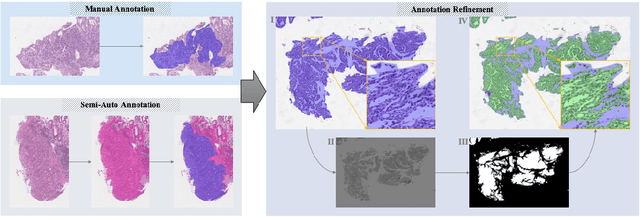
Abstract:Gastric cancer is one of the most common cancers, which ranks third among the leading causes of cancer death. Biopsy of gastric mucosal is a standard procedure in gastric cancer screening test. However, manual pathological inspection is labor-intensive and time-consuming. Besides, it is challenging for an automated algorithm to locate the small lesion regions in the gigapixel whole-slide image and make the decision correctly. To tackle these issues, we collected large-scale whole-slide image dataset with detailed lesion region annotation and designed a whole-slide image analyzing framework consisting of 3 networks which could not only determine the screen result but also present the suspicious areas to the pathologist for reference. Experiments demonstrated that our proposed framework achieves sensitivity of 97.05% and specificity of 92.72% in screening task and Dice coefficient of 0.8331 in segmentation task. Furthermore, we tested our best model in real-world scenario on 10, 316 whole-slide images collected from 4 medical centers.
Signet Ring Cell Detection With a Semi-supervised Learning Framework
Jul 09, 2019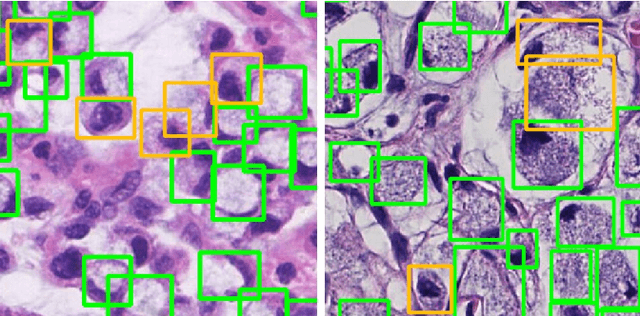


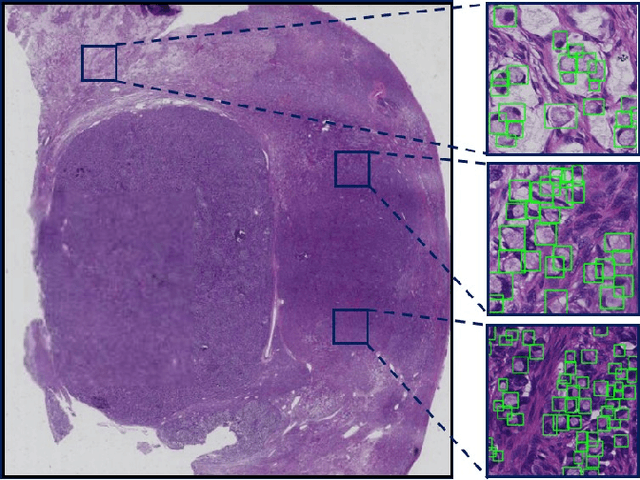
Abstract:Signet ring cell carcinoma is a type of rare adenocarcinoma with poor prognosis. Early detection leads to huge improvement of patients' survival rate. However, pathologists can only visually detect signet ring cells under the microscope. This procedure is not only laborious but also prone to omission. An automatic and accurate signet ring cell detection solution is thus important but has not been investigated before. In this paper, we take the first step to present a semi-supervised learning framework for the signet ring cell detection problem. Self-training is proposed to deal with the challenge of incomplete annotations, and cooperative-training is adapted to explore the unlabeled regions. Combining the two techniques, our semi-supervised learning framework can make better use of both labeled and unlabeled data. Experiments on large real clinical data demonstrate the effectiveness of our design. Our framework achieves accurate signet ring cell detection and can be readily applied in the clinical trails. The dataset will be released soon to facilitate the development of the area.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge