William Stafford Noble
GS-UW
Foundation model for mass spectrometry proteomics
May 19, 2025Abstract:Mass spectrometry is the dominant technology in the field of proteomics, enabling high-throughput analysis of the protein content of complex biological samples. Due to the complexity of the instrumentation and resulting data, sophisticated computational methods are required for the processing and interpretation of acquired mass spectra. Machine learning has shown great promise to improve the analysis of mass spectrometry data, with numerous purpose-built methods for improving specific steps in the data acquisition and analysis pipeline reaching widespread adoption. Here, we propose unifying various spectrum prediction tasks under a single foundation model for mass spectra. To this end, we pre-train a spectrum encoder using de novo sequencing as a pre-training task. We then show that using these pre-trained spectrum representations improves our performance on the four downstream tasks of spectrum quality prediction, chimericity prediction, phosphorylation prediction, and glycosylation status prediction. Finally, we perform multi-task fine-tuning and find that this approach improves the performance on each task individually. Overall, our work demonstrates that a foundation model for tandem mass spectrometry proteomics trained on de novo sequencing learns generalizable representations of spectra, improves performance on downstream tasks where training data is limited, and can ultimately enhance data acquisition and analysis in proteomics experiments.
Error-controlled non-additive interaction discovery in machine learning models
Aug 30, 2024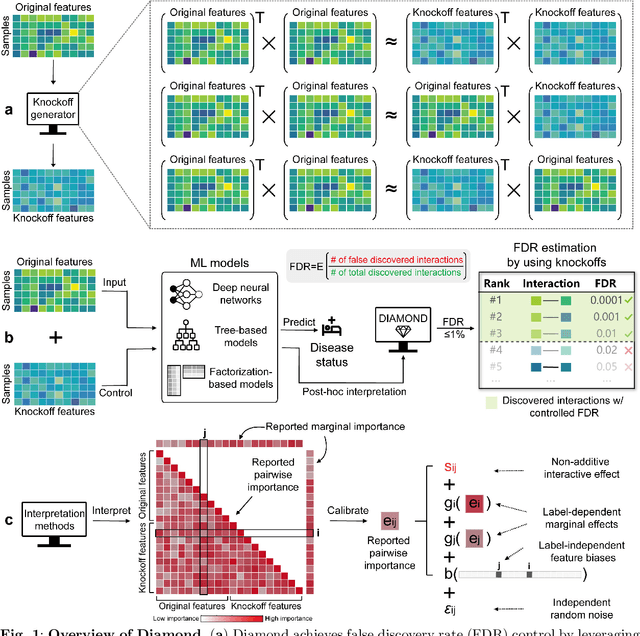
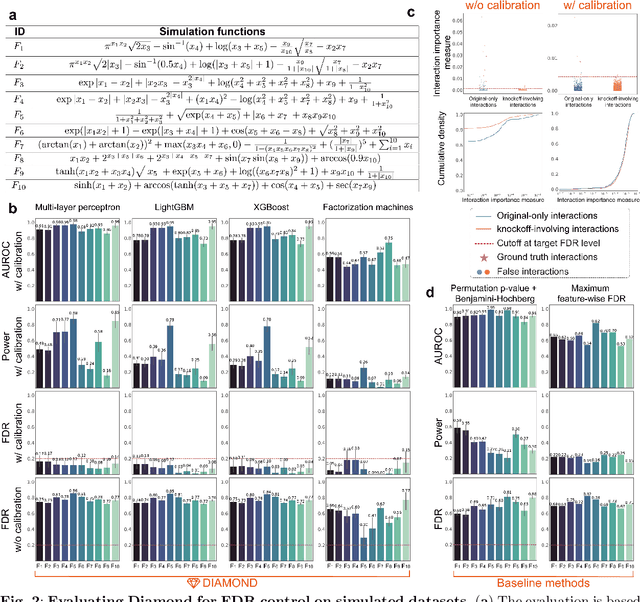
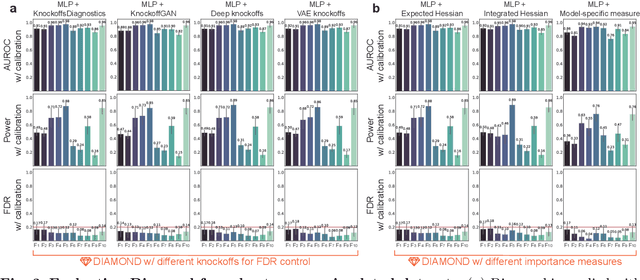
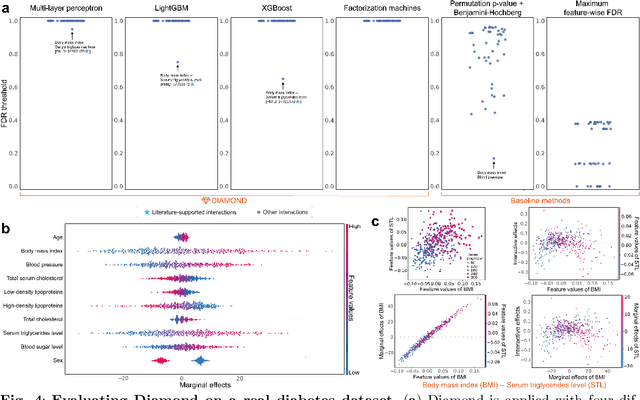
Abstract:Machine learning (ML) models are powerful tools for detecting complex patterns within data, yet their "black box" nature limits their interpretability, hindering their use in critical domains like healthcare and finance. To address this challenge, interpretable ML methods have been developed to explain how features influence model predictions. However, these methods often focus on univariate feature importance, overlooking the complex interactions between features that ML models are capable of capturing. Recognizing this limitation, recent efforts have aimed to extend these methods to discover feature interactions, but existing approaches struggle with robustness and error control, especially under data perturbations. In this study, we introduce Diamond, a novel method for trustworthy feature interaction discovery. Diamond uniquely integrates the model-X knockoffs framework to control the false discovery rate (FDR), ensuring that the proportion of falsely discovered interactions remains low. We further address the challenges of using off-the-shelf interaction importance measures by proposing a calibration procedure that refines these measures to maintain the desired FDR. Diamond's applicability spans a wide range of ML models, including deep neural networks, tree-based models, and factorization-based models. Our empirical evaluations on both simulated and real datasets across various biomedical studies demonstrate Diamond's utility in enabling more reliable data-driven scientific discoveries. This method represents a significant step forward in the deployment of ML models for scientific innovation and hypothesis generation.
DeepROCK: Error-controlled interaction detection in deep neural networks
Sep 26, 2023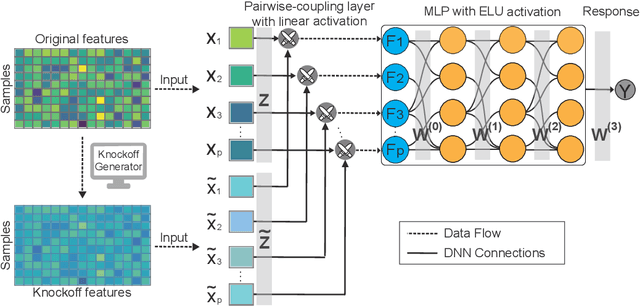
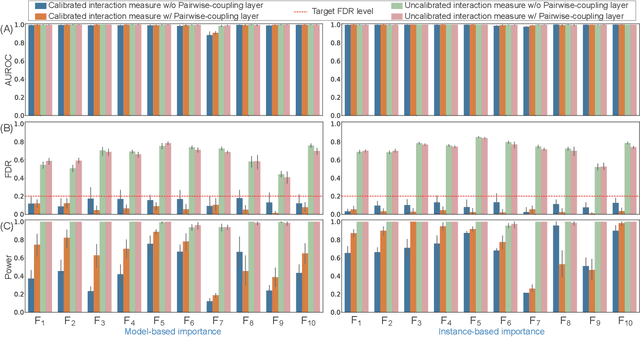
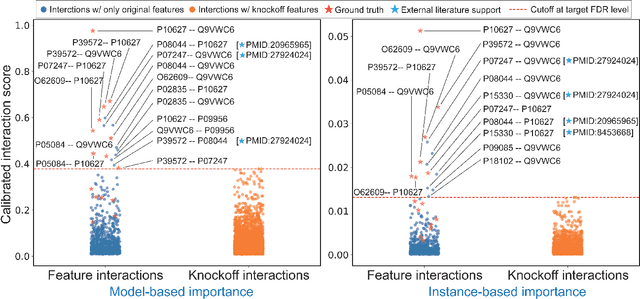
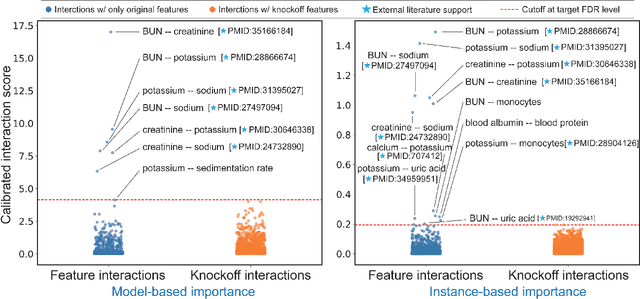
Abstract:The complexity of deep neural networks (DNNs) makes them powerful but also makes them challenging to interpret, hindering their applicability in error-intolerant domains. Existing methods attempt to reason about the internal mechanism of DNNs by identifying feature interactions that influence prediction outcomes. However, such methods typically lack a systematic strategy to prioritize interactions while controlling confidence levels, making them difficult to apply in practice for scientific discovery and hypothesis validation. In this paper, we introduce a method, called DeepROCK, to address this limitation by using knockoffs, which are dummy variables that are designed to mimic the dependence structure of a given set of features while being conditionally independent of the response. Together with a novel DNN architecture involving a pairwise-coupling layer, DeepROCK jointly controls the false discovery rate (FDR) and maximizes statistical power. In addition, we identify a challenge in correctly controlling FDR using off-the-shelf feature interaction importance measures. DeepROCK overcomes this challenge by proposing a calibration procedure applied to existing interaction importance measures to make the FDR under control at a target level. Finally, we validate the effectiveness of DeepROCK through extensive experiments on simulated and real datasets.
Robust saliency maps with decoy-enhanced saliency score
Feb 03, 2020



Abstract:Saliency methods help to make deep neural network predictions more interpretable by identifying particular features, such as pixels in an image, that contribute most strongly to the network's prediction. Unfortunately, recent evidence suggests that many saliency methods perform poorly when gradients are saturated or in the presence of strong inter-feature dependence or noise injected by an adversarial attack. In this work, we propose to infer robust saliency scores by integrating the saliency scores of a set of decoys with a novel decoy-enhanced saliency score, in which the decoys are generated by either solving an optimization problem or blurring the original input. We theoretically analyze that our method compensates for gradient saturation and considers joint activation patterns of pixels. We also apply our method to three different CNNs---VGGNet, AlexNet, and ResNet trained on ImageNet data set. The empirical results show both qualitatively and quantitatively that our method outperforms raw scores produced by three existing saliency methods, even in the presence of adversarial attacks.
apricot: Submodular selection for data summarization in Python
Jun 08, 2019
Abstract:We present apricot, an open source Python package for selecting representative subsets from large data sets using submodular optimization. The package implements an efficient greedy selection algorithm that offers strong theoretical guarantees on the quality of the selected set. Two submodular set functions are implemented in apricot: facility location, which is broadly applicable but requires memory quadratic in the number of examples in the data set, and a feature-based function that is less broadly applicable but can scale to millions of examples. Apricot is extremely efficient, using both algorithmic speedups such as the lazy greedy algorithm and code optimizers such as numba. We demonstrate the use of subset selection by training machine learning models to comparable accuracy using either the full data set or a representative subset thereof. This paper presents an explanation of submodular selection, an overview of the features in apricot, and an application to several data sets. The code and tutorial Jupyter notebooks are available at https://github.com/jmschrei/apricot
DeepPINK: reproducible feature selection in deep neural networks
Sep 06, 2018



Abstract:Deep learning has become increasingly popular in both supervised and unsupervised machine learning thanks to its outstanding empirical performance. However, because of their intrinsic complexity, most deep learning methods are largely treated as black box tools with little interpretability. Even though recent attempts have been made to facilitate the interpretability of deep neural networks (DNNs), existing methods are susceptible to noise and lack of robustness. Therefore, scientists are justifiably cautious about the reproducibility of the discoveries, which is often related to the interpretability of the underlying statistical models. In this paper, we describe a method to increase the interpretability and reproducibility of DNNs by incorporating the idea of feature selection with controlled error rate. By designing a new DNN architecture and integrating it with the recently proposed knockoffs framework, we perform feature selection with a controlled error rate, while maintaining high power. This new method, DeepPINK (Deep feature selection using Paired-Input Nonlinear Knockoffs), is applied to both simulated and real data sets to demonstrate its empirical utility.
Metric learning pairwise kernel for graph inference
Oct 21, 2006
Abstract:Much recent work in bioinformatics has focused on the inference of various types of biological networks, representing gene regulation, metabolic processes, protein-protein interactions, etc. A common setting involves inferring network edges in a supervised fashion from a set of high-confidence edges, possibly characterized by multiple, heterogeneous data sets (protein sequence, gene expression, etc.). Here, we distinguish between two modes of inference in this setting: direct inference based upon similarities between nodes joined by an edge, and indirect inference based upon similarities between one pair of nodes and another pair of nodes. We propose a supervised approach for the direct case by translating it into a distance metric learning problem. A relaxation of the resulting convex optimization problem leads to the support vector machine (SVM) algorithm with a particular kernel for pairs, which we call the metric learning pairwise kernel (MLPK). We demonstrate, using several real biological networks, that this direct approach often improves upon the state-of-the-art SVM for indirect inference with the tensor product pairwise kernel.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge