Jian Qiu
GS-UW
Leveraging Table Content for Zero-shot Text-to-SQL with Meta-Learning
Sep 12, 2021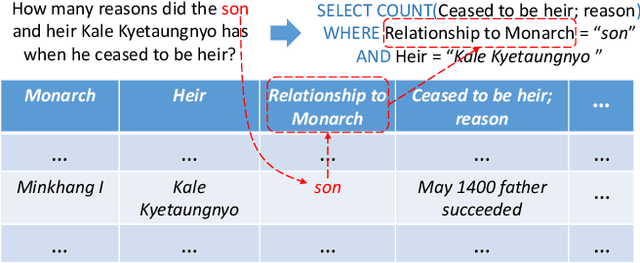
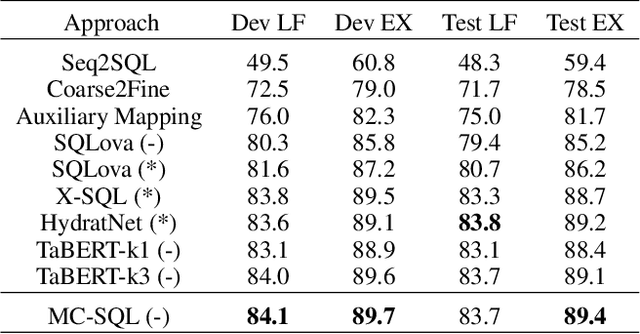
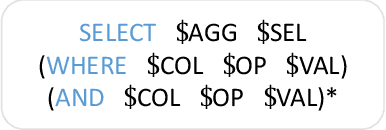
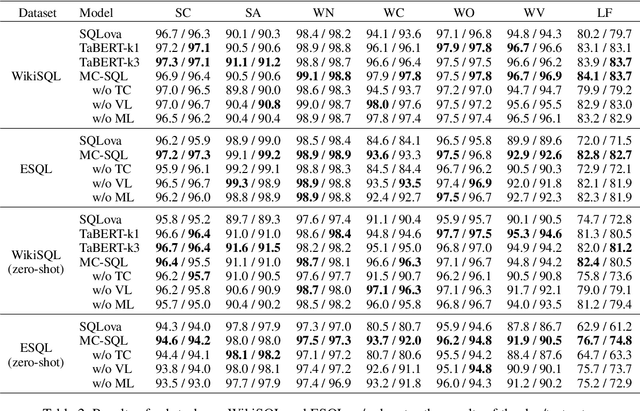
Abstract:Single-table text-to-SQL aims to transform a natural language question into a SQL query according to one single table. Recent work has made promising progress on this task by pre-trained language models and a multi-submodule framework. However, zero-shot table, that is, the invisible table in the training set, is currently the most critical bottleneck restricting the application of existing approaches to real-world scenarios. Although some work has utilized auxiliary tasks to help handle zero-shot tables, expensive extra manual annotation limits their practicality. In this paper, we propose a new approach for the zero-shot text-to-SQL task which does not rely on any additional manual annotations. Our approach consists of two parts. First, we propose a new model that leverages the abundant information of table content to help establish the mapping between questions and zero-shot tables. Further, we propose a simple but efficient meta-learning strategy to train our model. The strategy utilizes the two-step gradient update to force the model to learn a generalization ability towards zero-shot tables. We conduct extensive experiments on a public open-domain text-to-SQL dataset WikiSQL and a domain-specific dataset ESQL. Compared to existing approaches using the same pre-trained model, our approach achieves significant improvements on both datasets. Compared to the larger pre-trained model and the tabular-specific pre-trained model, our approach is still competitive. More importantly, on the zero-shot subsets of both the datasets, our approach further increases the improvements.
Metric learning pairwise kernel for graph inference
Oct 21, 2006
Abstract:Much recent work in bioinformatics has focused on the inference of various types of biological networks, representing gene regulation, metabolic processes, protein-protein interactions, etc. A common setting involves inferring network edges in a supervised fashion from a set of high-confidence edges, possibly characterized by multiple, heterogeneous data sets (protein sequence, gene expression, etc.). Here, we distinguish between two modes of inference in this setting: direct inference based upon similarities between nodes joined by an edge, and indirect inference based upon similarities between one pair of nodes and another pair of nodes. We propose a supervised approach for the direct case by translating it into a distance metric learning problem. A relaxation of the resulting convex optimization problem leads to the support vector machine (SVM) algorithm with a particular kernel for pairs, which we call the metric learning pairwise kernel (MLPK). We demonstrate, using several real biological networks, that this direct approach often improves upon the state-of-the-art SVM for indirect inference with the tensor product pairwise kernel.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge