Vinay Duddalwar
PSHop: A Lightweight Feed-Forward Method for 3D Prostate Gland Segmentation
Mar 24, 2024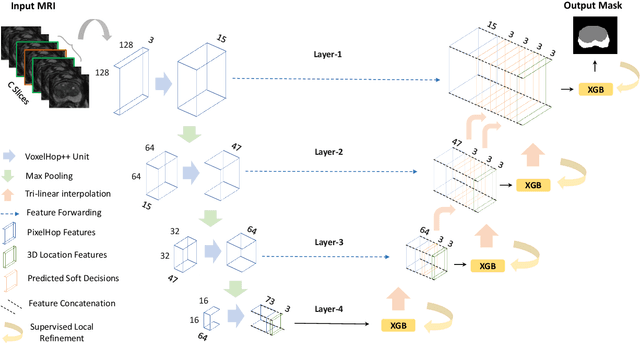

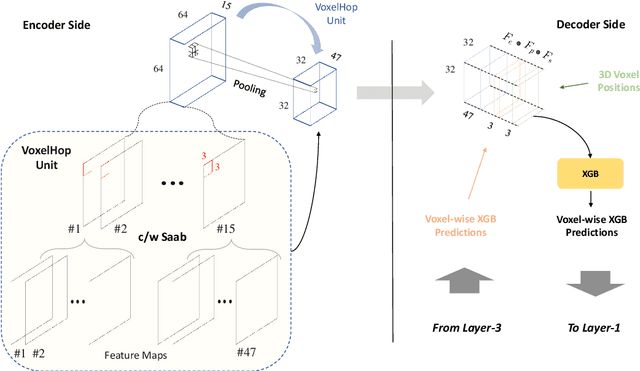
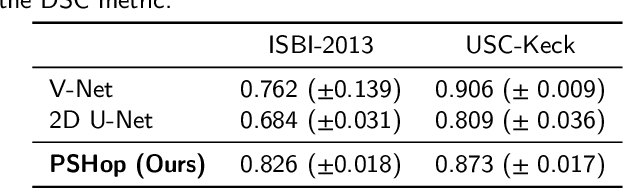
Abstract:Automatic prostate segmentation is an important step in computer-aided diagnosis of prostate cancer and treatment planning. Existing methods of prostate segmentation are based on deep learning models which have a large size and lack of transparency which is essential for physicians. In this paper, a new data-driven 3D prostate segmentation method on MRI is proposed, named PSHop. Different from deep learning based methods, the core methodology of PSHop is a feed-forward encoder-decoder system based on successive subspace learning (SSL). It consists of two modules: 1) encoder: fine to coarse unsupervised representation learning with cascaded VoxelHop units, 2) decoder: coarse to fine segmentation prediction with voxel-wise classification and local refinement. Experiments are conducted on the publicly available ISBI-2013 dataset, as well as on a larger private one. Experimental analysis shows that our proposed PSHop is effective, robust and lightweight in the tasks of prostate gland and zonal segmentation, achieving a Dice Similarity Coefficient (DSC) of 0.873 for the gland segmentation task. PSHop achieves a competitive performance comparatively to other deep learning methods, while keeping the model size and inference complexity an order of magnitude smaller.
PCa-RadHop: A Transparent and Lightweight Feed-forward Method for Clinically Significant Prostate Cancer Segmentation
Mar 24, 2024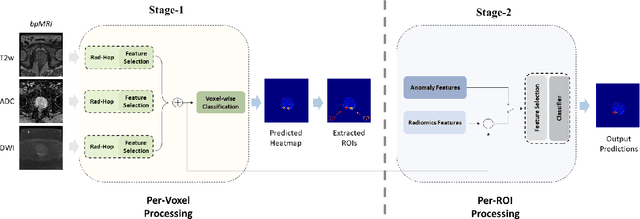


Abstract:Prostate Cancer is one of the most frequently occurring cancers in men, with a low survival rate if not early diagnosed. PI-RADS reading has a high false positive rate, thus increasing the diagnostic incurred costs and patient discomfort. Deep learning (DL) models achieve a high segmentation performance, although require a large model size and complexity. Also, DL models lack of feature interpretability and are perceived as ``black-boxes" in the medical field. PCa-RadHop pipeline is proposed in this work, aiming to provide a more transparent feature extraction process using a linear model. It adopts the recently introduced Green Learning (GL) paradigm, which offers a small model size and low complexity. PCa-RadHop consists of two stages: Stage-1 extracts data-driven radiomics features from the bi-parametric Magnetic Resonance Imaging (bp-MRI) input and predicts an initial heatmap. To reduce the false positive rate, a subsequent stage-2 is introduced to refine the predictions by including more contextual information and radiomics features from each already detected Region of Interest (ROI). Experiments on the largest publicly available dataset, PI-CAI, show a competitive performance standing of the proposed method among other deep DL models, achieving an area under the curve (AUC) of 0.807 among a cohort of 1,000 patients. Moreover, PCa-RadHop maintains orders of magnitude smaller model size and complexity.
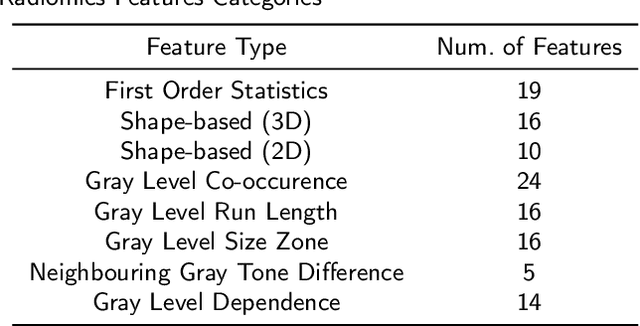
Benchmarking features from different radiomics toolkits / toolboxes using Image Biomarkers Standardization Initiative
Jun 23, 2020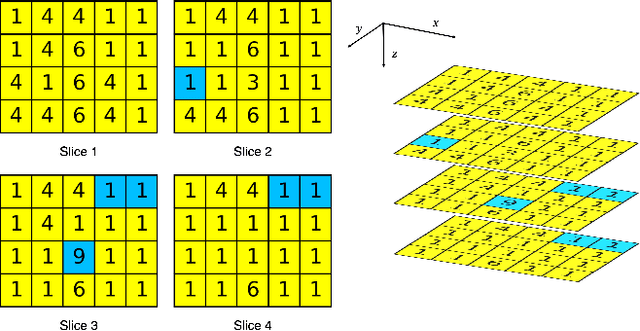
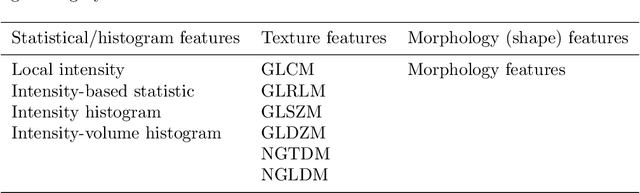
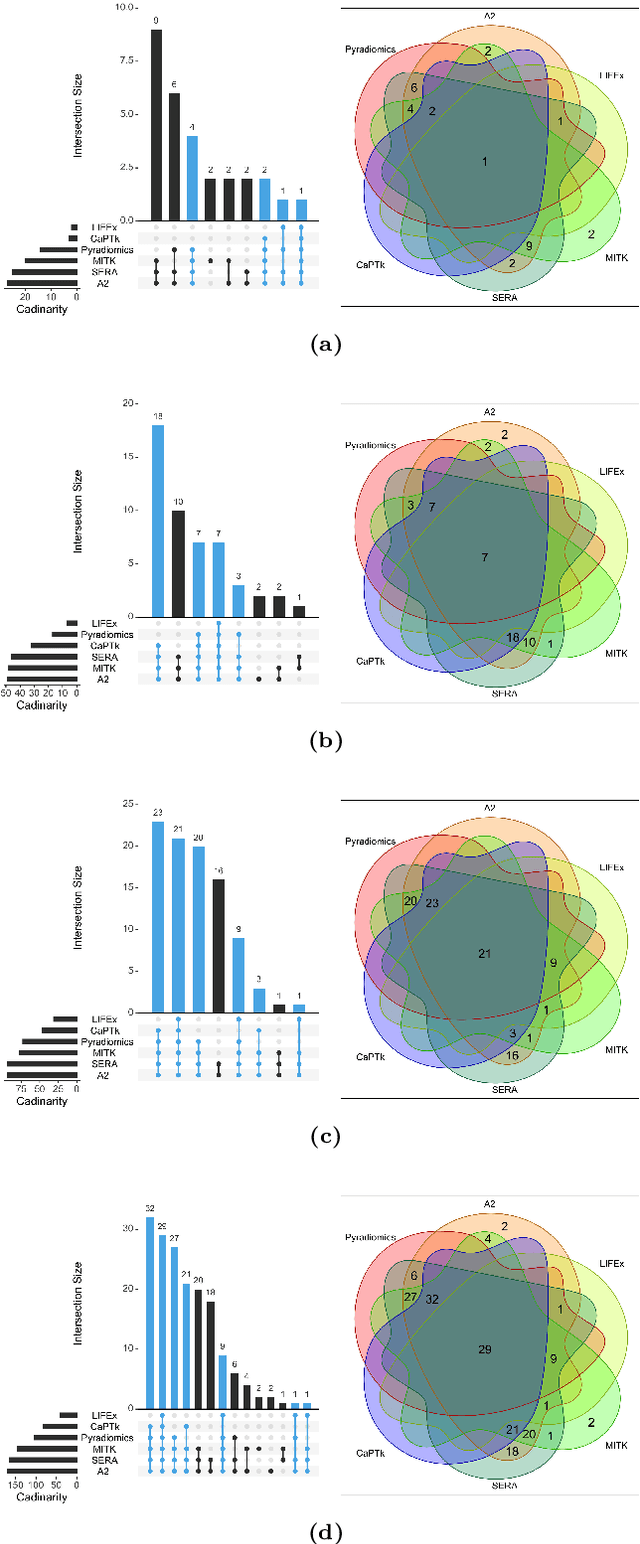
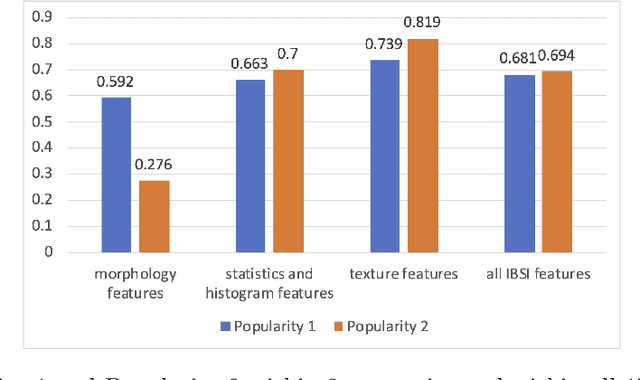
Abstract:There is no consensus regarding the radiomic feature terminology, the underlying mathematics, or their implementation. This creates a scenario where features extracted using different toolboxes could not be used to build or validate the same model leading to a non-generalization of radiomic results. In this study, the image biomarker standardization initiative (IBSI) established phantom and benchmark values were used to compare the variation of the radiomic features while using 6 publicly available software programs and 1 in-house radiomics pipeline. All IBSI-standardized features (11 classes, 173 in total) were extracted. The relative differences between the extracted feature values from the different software and the IBSI benchmark values were calculated to measure the inter-software agreement. To better understand the variations, features are further grouped into 3 categories according to their properties: 1) morphology, 2) statistic/histogram and 3)texture features. While a good agreement was observed for a majority of radiomics features across the various programs, relatively poor agreement was observed for morphology features. Significant differences were also found in programs that use different gray level discretization approaches. Since these programs do not include all IBSI features, the level of quantitative assessment for each category was analyzed using Venn and the UpSet diagrams and also quantified using two ad hoc metrics. Morphology features earns lowest scores for both metrics, indicating that morphological features are not consistently evaluated among software programs. We conclude that radiomic features calculated using different software programs may not be identical and reliable. Further studies are needed to standardize the workflow of radiomic feature extraction.
Role of Edge Device and Cloud Machine Learning in Point-of-Care Solutions Using Imaging Diagnostics for Population Screening
Jun 18, 2020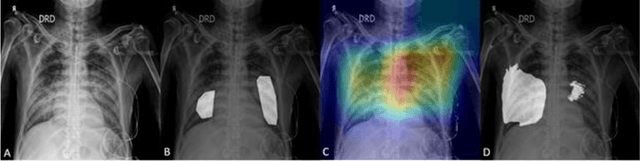
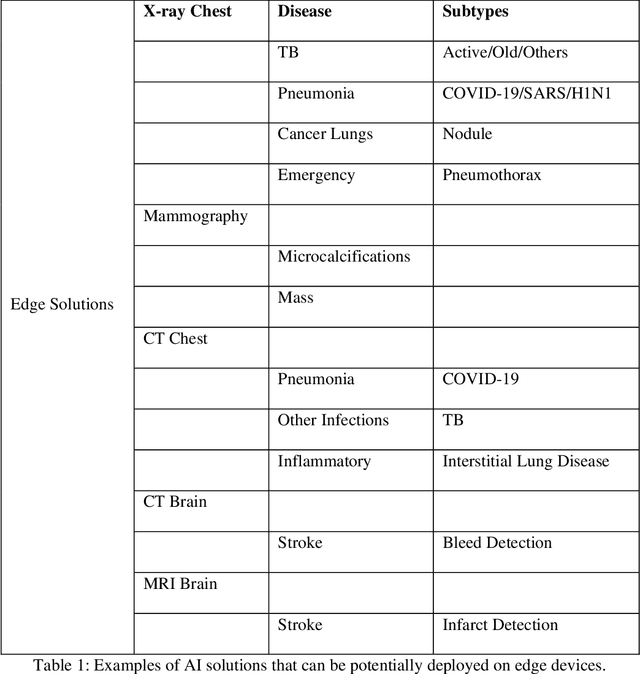
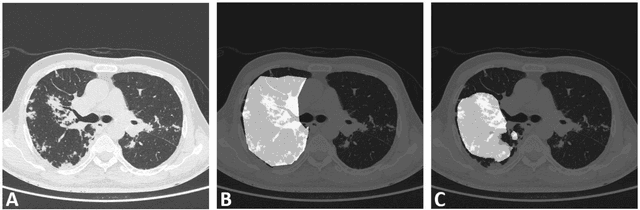
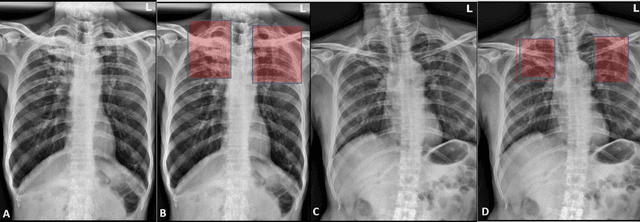
Abstract:Edge devices are revolutionizing diagnostics. Edge devices can reside within or adjacent to imaging tools such as digital Xray, CT, MRI, or ultrasound equipment. These devices are either CPUs or GPUs with advanced processing deep and machine learning (artificial intelligence) algorithms that assist in classification and triage solutions to flag studies as either normal or abnormal, TB or healthy (in case of TB screening), suspected COVID-19/other pneumonia or unremarkable (in hospital or hotspot settings). These can be deployed as screening point-of-care (PoC) solutions; this is particularly true for digital and portable X-ray devices. Edge device learning can also be used for mammography and CT studies where it can identify microcalcification and stroke, respectively. These solutions can be considered the first line of pre-screening before the imaging specialist actually reviews scans and makes a final diagnosis. The key advantage of these tools is that they are instant, can be deployed remotely where experts are not available to perform pre-screening before the experts actually review, and are not limited by internet bandwidth as the nano learning data centers are placed next to the device.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge