Viraj Kulkarni
Application of Federated Learning in Building a Robust COVID-19 Chest X-ray Classification Model
Apr 22, 2022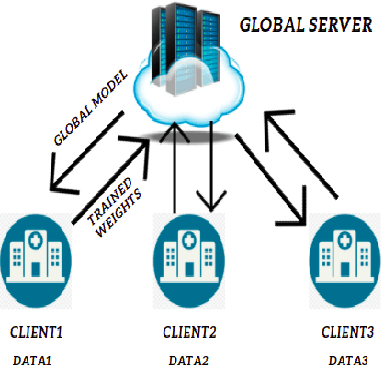
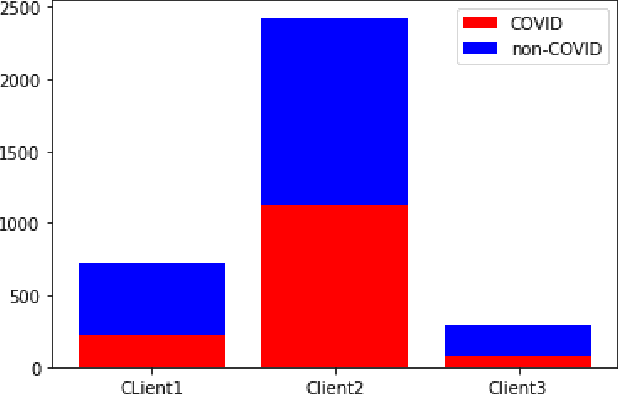
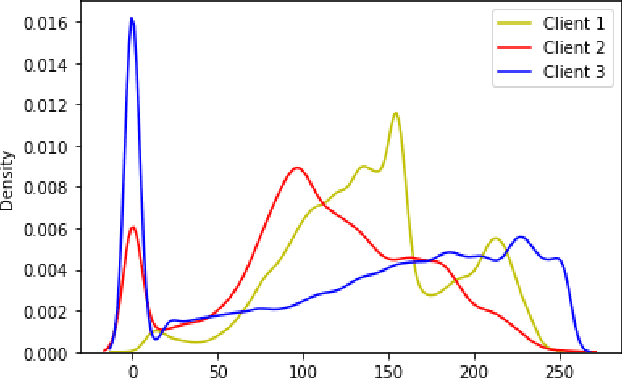
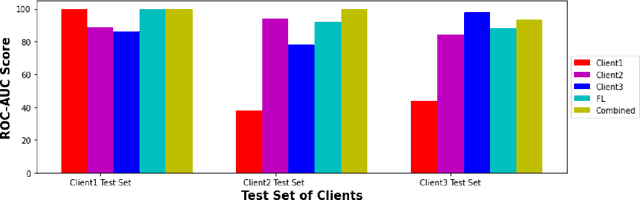
Abstract:While developing artificial intelligence (AI)-based algorithms to solve problems, the amount of data plays a pivotal role - large amount of data helps the researchers and engineers to develop robust AI algorithms. In the case of building AI-based models for problems related to medical imaging, these data need to be transferred from the medical institutions where they were acquired to the organizations developing the algorithms. This movement of data involves time-consuming formalities like complying with HIPAA, GDPR, etc.There is also a risk of patients' private data getting leaked, compromising their confidentiality. One solution to these problems is using the Federated Learning framework. Federated Learning (FL) helps AI models to generalize better and create a robust AI model by using data from different sources having different distributions and data characteristics without moving all the data to a central server. In our paper, we apply the FL framework for training a deep learning model to solve a binary classification problem of predicting the presence or absence of COVID-19. We took three different sources of data and trained individual models on each source. Then we trained an FL model on the complete data and compared all the model performances. We demonstrated that the FL model performs better than the individual models. Moreover, the FL model performed at par with the model trained on all the data combined at a central server. Thus Federated Learning leads to generalized AI models without the cost of data transfer and regulatory overhead.
A Classical-Quantum Convolutional Neural Network for Detecting Pneumonia from Chest Radiographs
Feb 19, 2022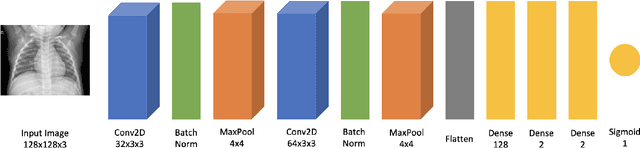

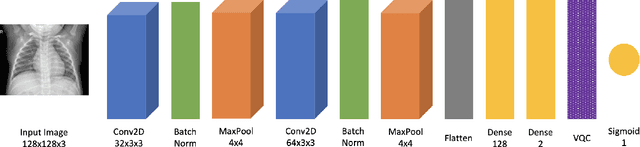
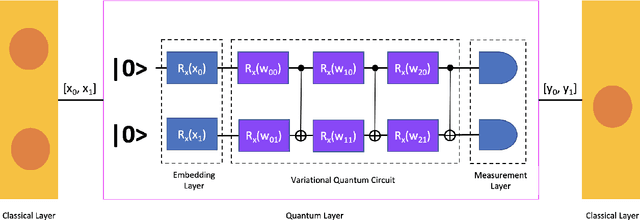
Abstract:While many quantum computing techniques for machine learning have been proposed, their performance on real-world datasets remains to be studied. In this paper, we explore how a variational quantum circuit could be integrated into a classical neural network for the problem of detecting pneumonia from chest radiographs. We substitute one layer of a classical convolutional neural network with a variational quantum circuit to create a hybrid neural network. We train both networks on an image dataset containing chest radiographs and benchmark their performance. To mitigate the influence of different sources of randomness in network training, we sample the results over multiple rounds. We show that the hybrid network outperforms the classical network on different performance measures, and that these improvements are statistically significant. Our work serves as an experimental demonstration of the potential of quantum computing to significantly improve neural network performance for real-world, non-trivial problems relevant to society and industry.
Levels of Autonomous Radiology
Dec 14, 2021Abstract:Radiology, being one of the younger disciplines of medicine with a history of just over a century, has witnessed tremendous technological advancements and has revolutionized the way we practice medicine today. In the last few decades, medical imaging modalities have generated seismic amounts of medical data. The development and adoption of Artificial Intelligence (AI) applications using this data will lead to the next phase of evolution in radiology. It will include automating laborious manual tasks such as annotations, report-generation, etc., along with the initial radiological assessment of cases to aid radiologists in their evaluation workflow. We propose a level-wise classification for the progression of automation in radiology, explaining AI assistance at each level with corresponding challenges and solutions. We hope that such discussions can help us address the challenges in a structured way and take the necessary steps to ensure the smooth adoption of new technologies in radiology.
Vulnerability Due to Training Order in Split Learning
Mar 26, 2021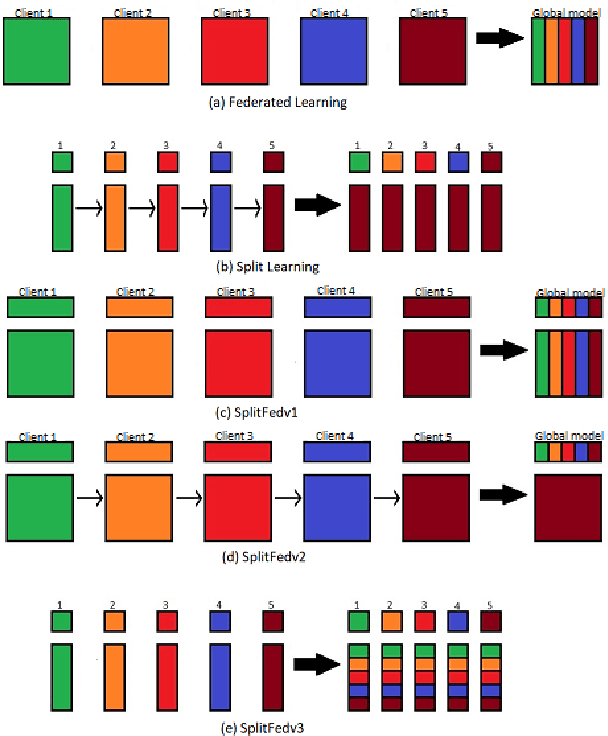
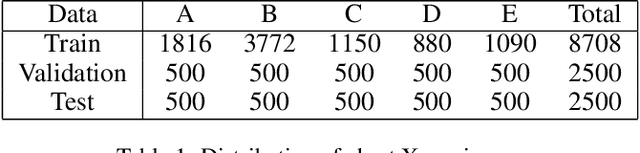
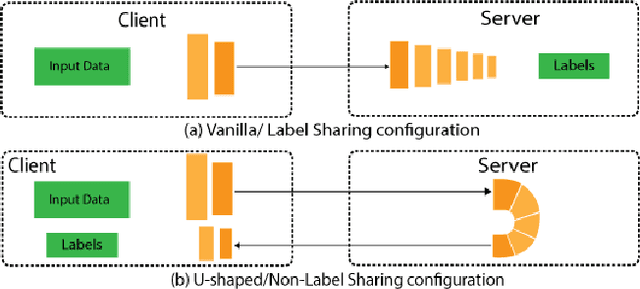
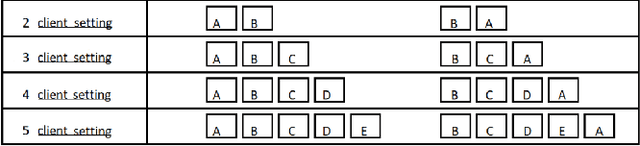
Abstract:Split learning (SL) is a privacy-preserving distributed deep learning method used to train a collaborative model without the need for sharing of patient's raw data between clients. In split learning, an additional privacy-preserving algorithm called no-peek algorithm can be incorporated, which is robust to adversarial attacks. The privacy benefits offered by split learning make it suitable for practice in the healthcare domain. However, the split learning algorithm is flawed as the collaborative model is trained sequentially, i.e., one client trains after the other. We point out that the model trained using the split learning algorithm gets biased towards the data of the clients used for training towards the end of a round. This makes SL algorithms highly susceptible to the order in which clients are considered for training. We demonstrate that the model trained using the data of all clients does not perform well on the client's data which was considered earliest in a round for training the model. Moreover, we show that this effect becomes more prominent with the increase in the number of clients. We also demonstrate that the SplitFedv3 algorithm mitigates this problem while still leveraging the privacy benefits provided by split learning.
Reducing Labelled Data Requirement for Pneumonia Segmentation using Image Augmentations
Feb 25, 2021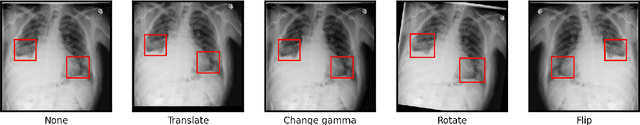
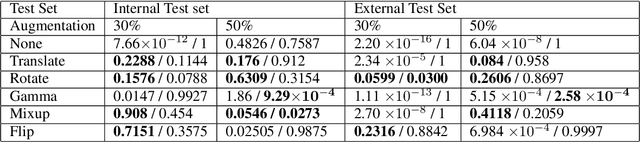
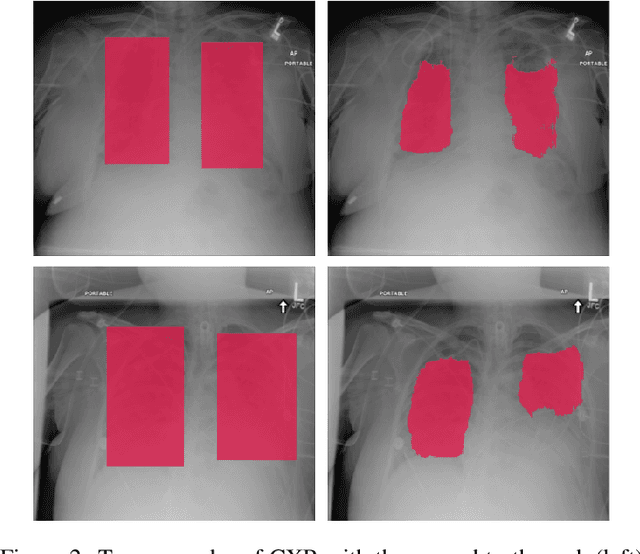
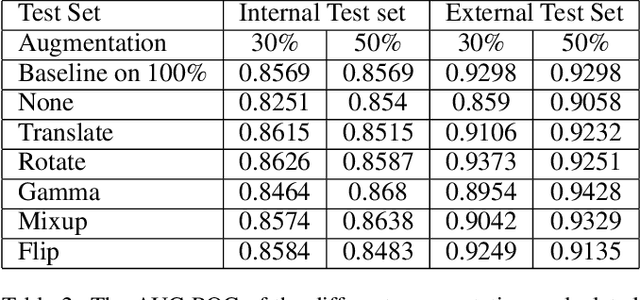
Abstract:Deep learning semantic segmentation algorithms can localise abnormalities or opacities from chest radiographs. However, the task of collecting and annotating training data is expensive and requires expertise which remains a bottleneck for algorithm performance. We investigate the effect of image augmentations on reducing the requirement of labelled data in the semantic segmentation of chest X-rays for pneumonia detection. We train fully convolutional network models on subsets of different sizes from the total training data. We apply a different image augmentation while training each model and compare it to the baseline trained on the entire dataset without augmentations. We find that rotate and mixup are the best augmentations amongst rotate, mixup, translate, gamma and horizontal flip, wherein they reduce the labelled data requirement by 70% while performing comparably to the baseline in terms of AUC and mean IoU in our experiments.
Key Technology Considerations in Developing and Deploying Machine Learning Models in Clinical Radiology Practice
Feb 03, 2021
Abstract:The use of machine learning to develop intelligent software tools for interpretation of radiology images has gained widespread attention in recent years. The development, deployment, and eventual adoption of these models in clinical practice, however, remains fraught with challenges. In this paper, we propose a list of key considerations that machine learning researchers must recognize and address to make their models accurate, robust, and usable in practice. Namely, we discuss: insufficient training data, decentralized datasets, high cost of annotations, ambiguous ground truth, imbalance in class representation, asymmetric misclassification costs, relevant performance metrics, generalization of models to unseen datasets, model decay, adversarial attacks, explainability, fairness and bias, and clinical validation. We describe each consideration and identify techniques to address it. Although these techniques have been discussed in prior research literature, by freshly examining them in the context of medical imaging and compiling them in the form of a laundry list, we hope to make them more accessible to researchers, software developers, radiologists, and other stakeholders.
Comparative Evaluation of 3D and 2D Deep Learning Techniques for Semantic Segmentation in CT Scans
Jan 19, 2021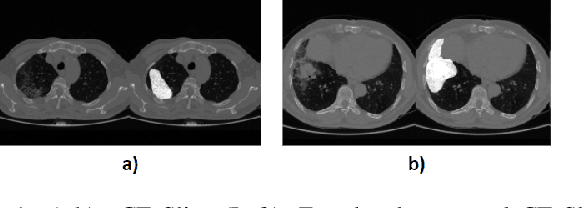
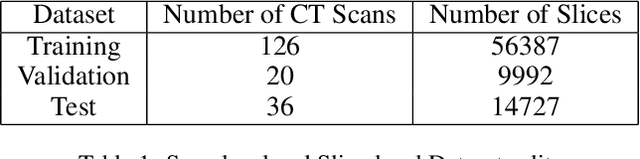
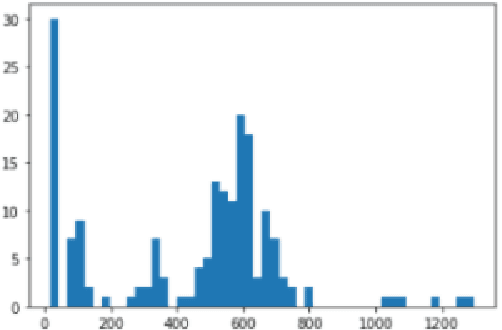
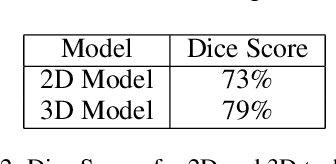
Abstract:Image segmentation plays a pivotal role in several medical-imaging applications by assisting the segmentation of the regions of interest. Deep learning-based approaches have been widely adopted for semantic segmentation of medical data. In recent years, in addition to 2D deep learning architectures, 3D architectures have been employed as the predictive algorithms for 3D medical image data. In this paper, we propose a 3D stack-based deep learning technique for segmenting manifestations of consolidation and ground-glass opacities in 3D Computed Tomography (CT) scans. We also present a comparison based on the segmentation results, the contextual information retained, and the inference time between this 3D technique and a traditional 2D deep learning technique. We also define the area-plot, which represents the peculiar pattern observed in the slice-wise areas of the pathology regions predicted by these deep learning models. In our exhaustive evaluation, 3D technique performs better than the 2D technique for the segmentation of CT scans. We get dice scores of 79% and 73% for the 3D and the 2D techniques respectively. The 3D technique results in a 5X reduction in the inference time compared to the 2D technique. Results also show that the area-plots predicted by the 3D model are more similar to the ground truth than those predicted by the 2D model. We also show how increasing the amount of contextual information retained during the training can improve the 3D model's performance.
Deep Learning Models for Calculation of Cardiothoracic Ratio from Chest Radiographs for Assisted Diagnosis of Cardiomegaly
Jan 19, 2021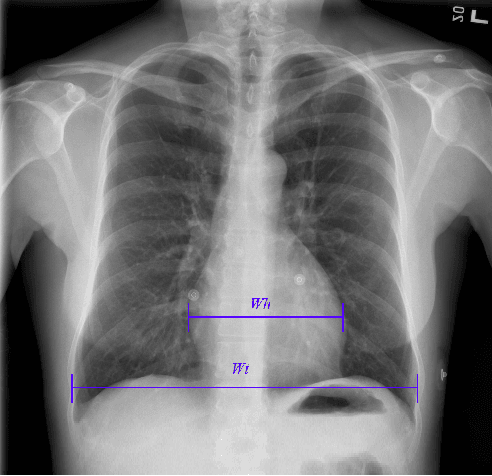
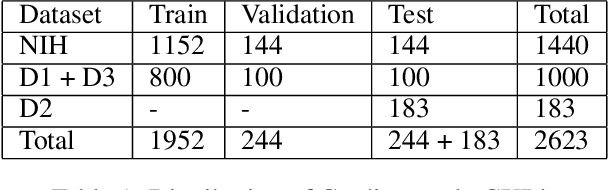
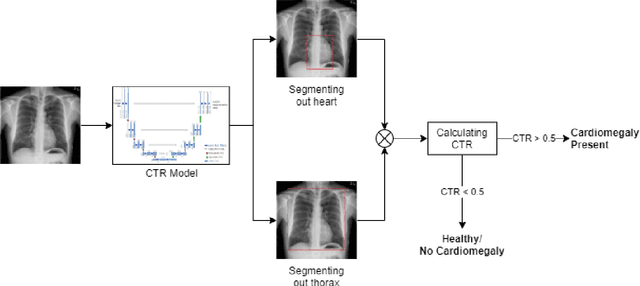
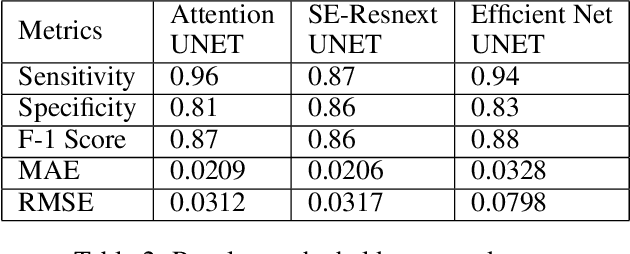
Abstract:We propose an automated method based on deep learning to compute the cardiothoracic ratio and detect the presence of cardiomegaly from chest radiographs. We develop two separate models to demarcate the heart and chest regions in an X-ray image using bounding boxes and use their outputs to calculate the cardiothoracic ratio. We obtain a sensitivity of 0.96 at a specificity of 0.81 with a mean absolute error of 0.0209 on a held-out test dataset and a sensitivity of 0.84 at a specificity of 0.97 with a mean absolute error of 0.018 on an independent dataset from a different hospital. We also compare three different segmentation model architectures for the proposed method and observe that Attention U-Net yields better results than SE-Resnext U-Net and EfficientNet U-Net. By providing a numeric measurement of the cardiothoracic ratio, we hope to mitigate human subjectivity arising out of visual assessment in the detection of cardiomegaly.
Comparison of Privacy-Preserving Distributed Deep Learning Methods in Healthcare
Dec 23, 2020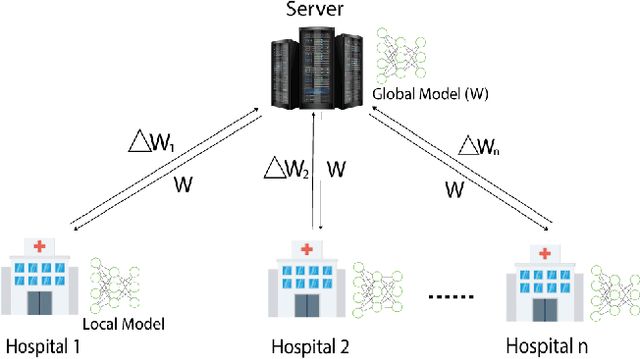
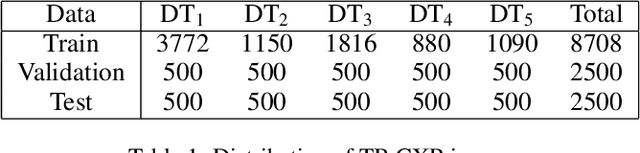
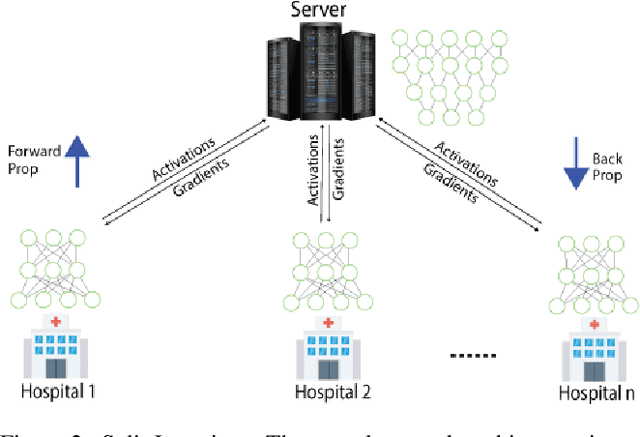
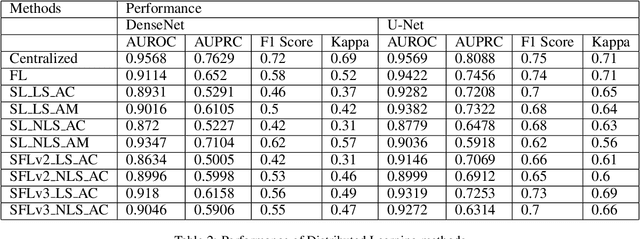
Abstract:In this paper, we compare three privacy-preserving distributed learning techniques: federated learning, split learning, and SplitFed. We use these techniques to develop binary classification models for detecting tuberculosis from chest X-rays and compare them in terms of classification performance, communication and computational costs, and training time. We propose a novel distributed learning architecture called SplitFedv3, which performs better than split learning and SplitFedv2 in our experiments. We also propose alternate mini-batch training, a new training technique for split learning, that performs better than alternate client training, where clients take turns to train a model.
Automated Detection of COVID-19 from CT Scans Using Convolutional Neural Networks
Jun 23, 2020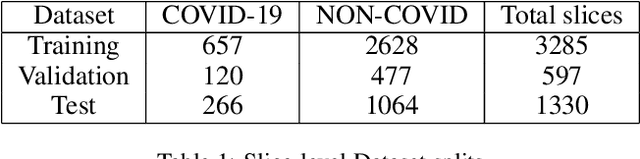
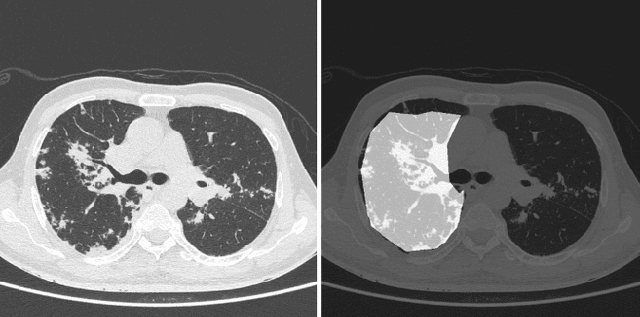
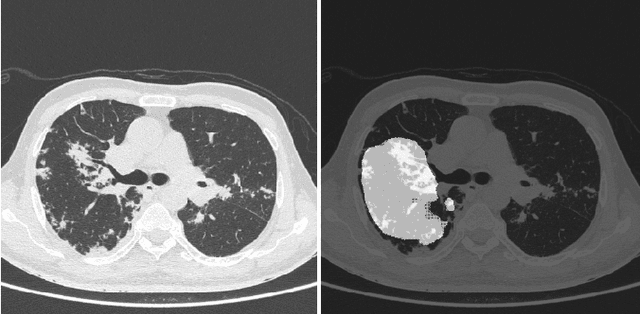
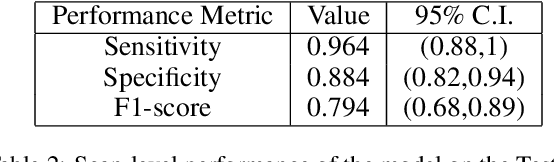
Abstract:COVID-19 is an infectious disease that causes respiratory problems similar to those caused by SARS-CoV (2003). Currently, swab samples are being used for its diagnosis. The most common testing method used is the RT-PCR method, which has high specificity but variable sensitivity. AI-based detection has the capability to overcome this drawback. In this paper, we propose a prospective method wherein we use chest CT scans to diagnose the patients for COVID-19 pneumonia. We use a set of open-source images, available as individual CT slices, and full CT scans from a private Indian Hospital to train our model. We build a 2D segmentation model using the U-Net architecture, which gives the output by marking out the region of infection. Our model achieves a sensitivity of 96.428% (95% CI: 88%-100%) and a specificity of 88.39% (95% CI: 82%-94%). Additionally, we derive a logic for converting our slice-level predictions to scan-level, which helps us reduce the false positives.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge