Giovanni Cacciamani
PSHop: A Lightweight Feed-Forward Method for 3D Prostate Gland Segmentation
Mar 24, 2024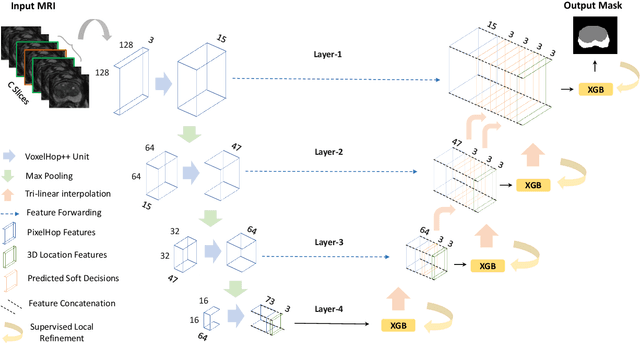

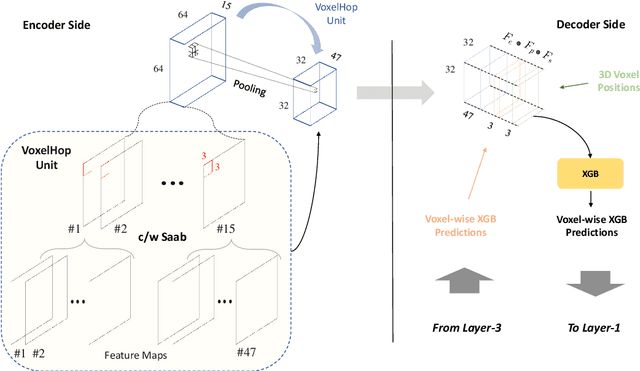
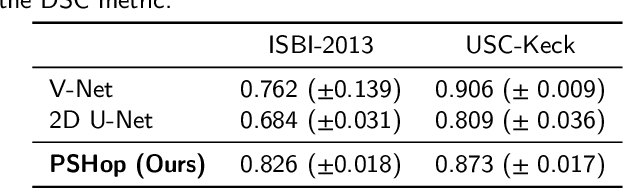
Abstract:Automatic prostate segmentation is an important step in computer-aided diagnosis of prostate cancer and treatment planning. Existing methods of prostate segmentation are based on deep learning models which have a large size and lack of transparency which is essential for physicians. In this paper, a new data-driven 3D prostate segmentation method on MRI is proposed, named PSHop. Different from deep learning based methods, the core methodology of PSHop is a feed-forward encoder-decoder system based on successive subspace learning (SSL). It consists of two modules: 1) encoder: fine to coarse unsupervised representation learning with cascaded VoxelHop units, 2) decoder: coarse to fine segmentation prediction with voxel-wise classification and local refinement. Experiments are conducted on the publicly available ISBI-2013 dataset, as well as on a larger private one. Experimental analysis shows that our proposed PSHop is effective, robust and lightweight in the tasks of prostate gland and zonal segmentation, achieving a Dice Similarity Coefficient (DSC) of 0.873 for the gland segmentation task. PSHop achieves a competitive performance comparatively to other deep learning methods, while keeping the model size and inference complexity an order of magnitude smaller.
PCa-RadHop: A Transparent and Lightweight Feed-forward Method for Clinically Significant Prostate Cancer Segmentation
Mar 24, 2024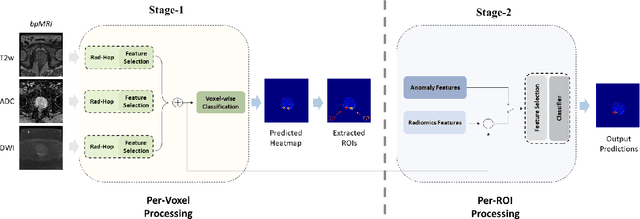


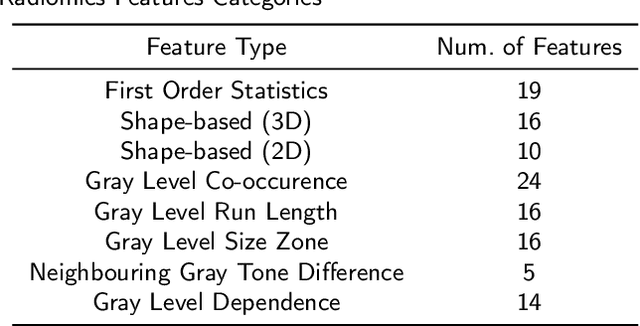
Abstract:Prostate Cancer is one of the most frequently occurring cancers in men, with a low survival rate if not early diagnosed. PI-RADS reading has a high false positive rate, thus increasing the diagnostic incurred costs and patient discomfort. Deep learning (DL) models achieve a high segmentation performance, although require a large model size and complexity. Also, DL models lack of feature interpretability and are perceived as ``black-boxes" in the medical field. PCa-RadHop pipeline is proposed in this work, aiming to provide a more transparent feature extraction process using a linear model. It adopts the recently introduced Green Learning (GL) paradigm, which offers a small model size and low complexity. PCa-RadHop consists of two stages: Stage-1 extracts data-driven radiomics features from the bi-parametric Magnetic Resonance Imaging (bp-MRI) input and predicts an initial heatmap. To reduce the false positive rate, a subsequent stage-2 is introduced to refine the predictions by including more contextual information and radiomics features from each already detected Region of Interest (ROI). Experiments on the largest publicly available dataset, PI-CAI, show a competitive performance standing of the proposed method among other deep DL models, achieving an area under the curve (AUC) of 0.807 among a cohort of 1,000 patients. Moreover, PCa-RadHop maintains orders of magnitude smaller model size and complexity.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge