Inderbir S. Gill
PSHop: A Lightweight Feed-Forward Method for 3D Prostate Gland Segmentation
Mar 24, 2024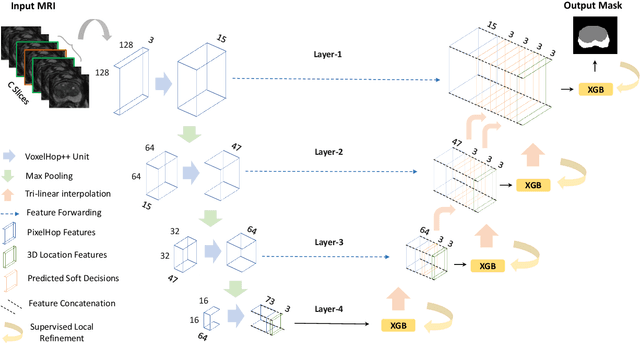

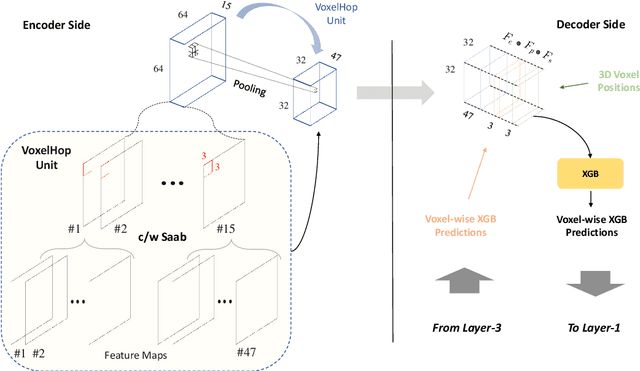
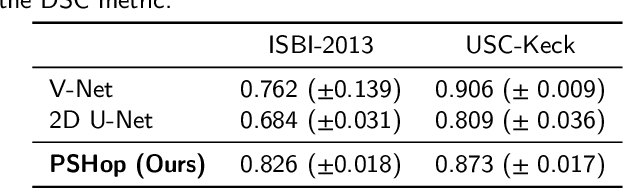
Abstract:Automatic prostate segmentation is an important step in computer-aided diagnosis of prostate cancer and treatment planning. Existing methods of prostate segmentation are based on deep learning models which have a large size and lack of transparency which is essential for physicians. In this paper, a new data-driven 3D prostate segmentation method on MRI is proposed, named PSHop. Different from deep learning based methods, the core methodology of PSHop is a feed-forward encoder-decoder system based on successive subspace learning (SSL). It consists of two modules: 1) encoder: fine to coarse unsupervised representation learning with cascaded VoxelHop units, 2) decoder: coarse to fine segmentation prediction with voxel-wise classification and local refinement. Experiments are conducted on the publicly available ISBI-2013 dataset, as well as on a larger private one. Experimental analysis shows that our proposed PSHop is effective, robust and lightweight in the tasks of prostate gland and zonal segmentation, achieving a Dice Similarity Coefficient (DSC) of 0.873 for the gland segmentation task. PSHop achieves a competitive performance comparatively to other deep learning methods, while keeping the model size and inference complexity an order of magnitude smaller.
PCa-RadHop: A Transparent and Lightweight Feed-forward Method for Clinically Significant Prostate Cancer Segmentation
Mar 24, 2024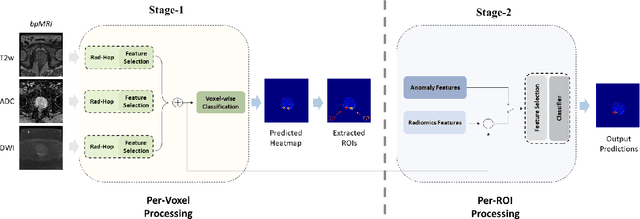


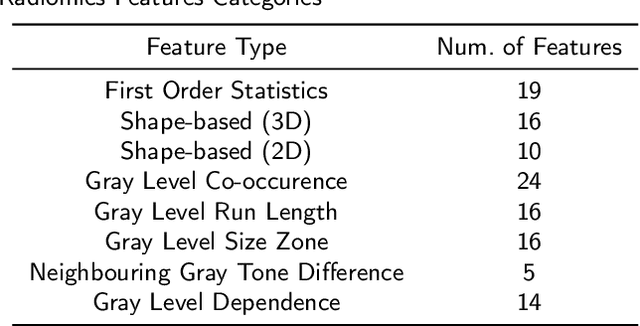
Abstract:Prostate Cancer is one of the most frequently occurring cancers in men, with a low survival rate if not early diagnosed. PI-RADS reading has a high false positive rate, thus increasing the diagnostic incurred costs and patient discomfort. Deep learning (DL) models achieve a high segmentation performance, although require a large model size and complexity. Also, DL models lack of feature interpretability and are perceived as ``black-boxes" in the medical field. PCa-RadHop pipeline is proposed in this work, aiming to provide a more transparent feature extraction process using a linear model. It adopts the recently introduced Green Learning (GL) paradigm, which offers a small model size and low complexity. PCa-RadHop consists of two stages: Stage-1 extracts data-driven radiomics features from the bi-parametric Magnetic Resonance Imaging (bp-MRI) input and predicts an initial heatmap. To reduce the false positive rate, a subsequent stage-2 is introduced to refine the predictions by including more contextual information and radiomics features from each already detected Region of Interest (ROI). Experiments on the largest publicly available dataset, PI-CAI, show a competitive performance standing of the proposed method among other deep DL models, achieving an area under the curve (AUC) of 0.807 among a cohort of 1,000 patients. Moreover, PCa-RadHop maintains orders of magnitude smaller model size and complexity.
Bibliometric Analysis of Publisher and Journal Instructions to Authors on Generative-AI in Academic and Scientific Publishing
Jul 21, 2023Abstract:We aim to determine the extent and content of guidance for authors regarding the use of generative-AI (GAI), Generative Pretrained models (GPTs) and Large Language Models (LLMs) powered tools among the top 100 academic publishers and journals in science. The websites of these publishers and journals were screened from between 19th and 20th May 2023. Among the largest 100 publishers, 17% provided guidance on the use of GAI, of which 12 (70.6%) were among the top 25 publishers. Among the top 100 journals, 70% have provided guidance on GAI. Of those with guidance, 94.1% of publishers and 95.7% of journals prohibited the inclusion of GAI as an author. Four journals (5.7%) explicitly prohibit the use of GAI in the generation of a manuscript, while 3 (17.6%) publishers and 15 (21.4%) journals indicated their guidance exclusively applies to the writing process. When disclosing the use of GAI, 42.8% of publishers and 44.3% of journals included specific disclosure criteria. There was variability in guidance of where to disclose the use of GAI, including in the methods, acknowledgments, cover letter, or a new section. There was also variability in how to access GAI guidance and the linking of journal and publisher instructions to authors. There is a lack of guidance by some top publishers and journals on the use of GAI by authors. Among those publishers and journals that provide guidance, there is substantial heterogeneity in the allowable uses of GAI and in how it should be disclosed, with this heterogeneity persisting among affiliated publishers and journals in some instances. The lack of standardization burdens authors and threatens to limit the effectiveness of these regulations. There is a need for standardized guidelines in order to protect the integrity of scientific output as GAI continues to grow in popularity.
Development of the ChatGPT, Generative Artificial Intelligence and Natural Large Language Models for Accountable Reporting and Use (CANGARU) Guidelines
Jul 18, 2023
Abstract:The swift progress and ubiquitous adoption of Generative AI (GAI), Generative Pre-trained Transformers (GPTs), and large language models (LLMs) like ChatGPT, have spurred queries about their ethical application, use, and disclosure in scholarly research and scientific productions. A few publishers and journals have recently created their own sets of rules; however, the absence of a unified approach may lead to a 'Babel Tower Effect,' potentially resulting in confusion rather than desired standardization. In response to this, we present the ChatGPT, Generative Artificial Intelligence, and Natural Large Language Models for Accountable Reporting and Use Guidelines (CANGARU) initiative, with the aim of fostering a cross-disciplinary global inclusive consensus on the ethical use, disclosure, and proper reporting of GAI/GPT/LLM technologies in academia. The present protocol consists of four distinct parts: a) an ongoing systematic review of GAI/GPT/LLM applications to understand the linked ideas, findings, and reporting standards in scholarly research, and to formulate guidelines for its use and disclosure, b) a bibliometric analysis of existing author guidelines in journals that mention GAI/GPT/LLM, with the goal of evaluating existing guidelines, analyzing the disparity in their recommendations, and identifying common rules that can be brought into the Delphi consensus process, c) a Delphi survey to establish agreement on the items for the guidelines, ensuring principled GAI/GPT/LLM use, disclosure, and reporting in academia, and d) the subsequent development and dissemination of the finalized guidelines and their supplementary explanation and elaboration documents.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge