Vasileios Magoulianitis
LG-NuSegHop: A Local-to-Global Self-Supervised Pipeline For Nuclei Instance Segmentation
Nov 07, 2025Abstract:Nuclei segmentation is the cornerstone task in histology image reading, shedding light on the underlying molecular patterns and leading to disease or cancer diagnosis. Yet, it is a laborious task that requires expertise from trained physicians. The large nuclei variability across different organ tissues and acquisition processes challenges the automation of this task. On the other hand, data annotations are expensive to obtain, and thus, Deep Learning (DL) models are challenged to generalize to unseen organs or different domains. This work proposes Local-to-Global NuSegHop (LG-NuSegHop), a self-supervised pipeline developed on prior knowledge of the problem and molecular biology. There are three distinct modules: (1) a set of local processing operations to generate a pseudolabel, (2) NuSegHop a novel data-driven feature extraction model and (3) a set of global operations to post-process the predictions of NuSegHop. Notably, even though the proposed pipeline uses { no manually annotated training data} or domain adaptation, it maintains a good generalization performance on other datasets. Experiments in three publicly available datasets show that our method outperforms other self-supervised and weakly supervised methods while having a competitive standing among fully supervised methods. Remarkably, every module within LG-NuSegHop is transparent and explainable to physicians.
* 42 pages, 8 figures, 7 tables
RadHop-Net: A Lightweight Radiomics-to-Error Regression for False Positive Reduction In MRI Prostate Cancer Detection
Jan 03, 2025



Abstract:Clinically significant prostate cancer (csPCa) is a leading cause of cancer death in men, yet it has a high survival rate if diagnosed early. Bi-parametric MRI (bpMRI) reading has become a prominent screening test for csPCa. However, this process has a high false positive (FP) rate, incurring higher diagnostic costs and patient discomfort. This paper introduces RadHop-Net, a novel and lightweight CNN for FP reduction. The pipeline consists of two stages: Stage 1 employs data driven radiomics to extract candidate ROIs. In contrast, Stage 2 expands the receptive field about each ROI using RadHop-Net to compensate for the predicted error from Stage 1. Moreover, a novel loss function for regression problems is introduced to balance the influence between FPs and true positives (TPs). RadHop-Net is trained in a radiomics-to-error manner, thus decoupling from the common voxel-to-label approach. The proposed Stage 2 improves the average precision (AP) in lesion detection from 0.407 to 0.468 in the publicly available pi-cai dataset, also maintaining a significantly smaller model size than the state-of-the-art.
PCa-RadHop: A Transparent and Lightweight Feed-forward Method for Clinically Significant Prostate Cancer Segmentation
Mar 24, 2024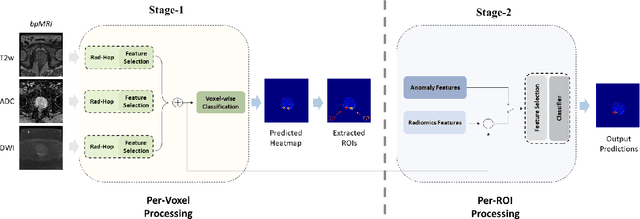


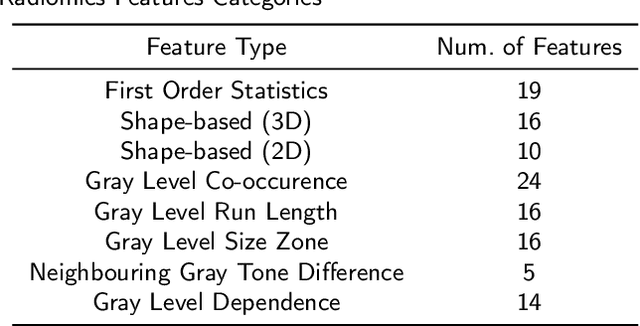
Abstract:Prostate Cancer is one of the most frequently occurring cancers in men, with a low survival rate if not early diagnosed. PI-RADS reading has a high false positive rate, thus increasing the diagnostic incurred costs and patient discomfort. Deep learning (DL) models achieve a high segmentation performance, although require a large model size and complexity. Also, DL models lack of feature interpretability and are perceived as ``black-boxes" in the medical field. PCa-RadHop pipeline is proposed in this work, aiming to provide a more transparent feature extraction process using a linear model. It adopts the recently introduced Green Learning (GL) paradigm, which offers a small model size and low complexity. PCa-RadHop consists of two stages: Stage-1 extracts data-driven radiomics features from the bi-parametric Magnetic Resonance Imaging (bp-MRI) input and predicts an initial heatmap. To reduce the false positive rate, a subsequent stage-2 is introduced to refine the predictions by including more contextual information and radiomics features from each already detected Region of Interest (ROI). Experiments on the largest publicly available dataset, PI-CAI, show a competitive performance standing of the proposed method among other deep DL models, achieving an area under the curve (AUC) of 0.807 among a cohort of 1,000 patients. Moreover, PCa-RadHop maintains orders of magnitude smaller model size and complexity.
PSHop: A Lightweight Feed-Forward Method for 3D Prostate Gland Segmentation
Mar 24, 2024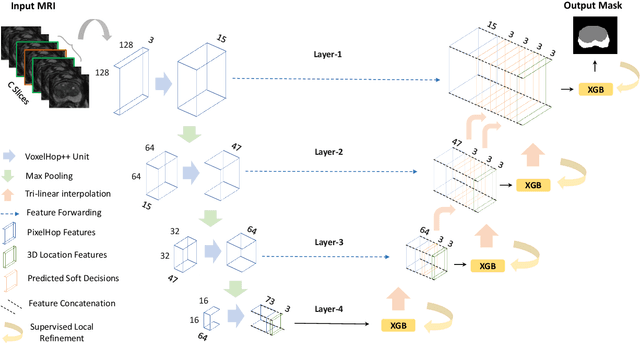

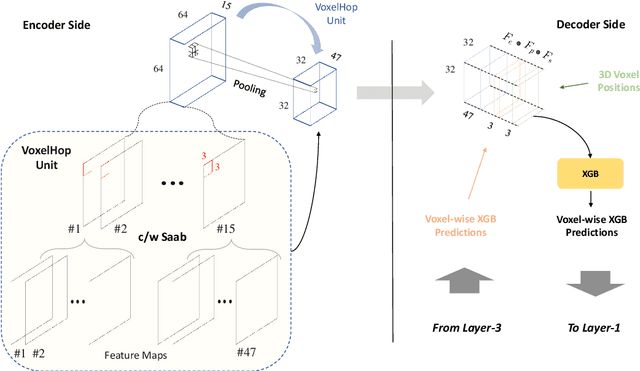
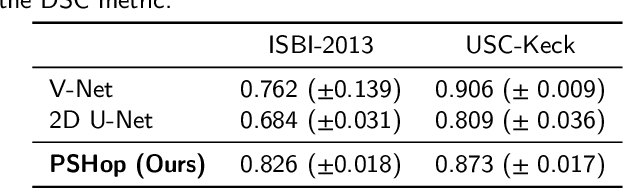
Abstract:Automatic prostate segmentation is an important step in computer-aided diagnosis of prostate cancer and treatment planning. Existing methods of prostate segmentation are based on deep learning models which have a large size and lack of transparency which is essential for physicians. In this paper, a new data-driven 3D prostate segmentation method on MRI is proposed, named PSHop. Different from deep learning based methods, the core methodology of PSHop is a feed-forward encoder-decoder system based on successive subspace learning (SSL). It consists of two modules: 1) encoder: fine to coarse unsupervised representation learning with cascaded VoxelHop units, 2) decoder: coarse to fine segmentation prediction with voxel-wise classification and local refinement. Experiments are conducted on the publicly available ISBI-2013 dataset, as well as on a larger private one. Experimental analysis shows that our proposed PSHop is effective, robust and lightweight in the tasks of prostate gland and zonal segmentation, achieving a Dice Similarity Coefficient (DSC) of 0.873 for the gland segmentation task. PSHop achieves a competitive performance comparatively to other deep learning methods, while keeping the model size and inference complexity an order of magnitude smaller.
A Comprehensive Overview of Computational Nuclei Segmentation Methods in Digital Pathology
Aug 16, 2023Abstract:In the cancer diagnosis pipeline, digital pathology plays an instrumental role in the identification, staging, and grading of malignant areas on biopsy tissue specimens. High resolution histology images are subject to high variance in appearance, sourcing either from the acquisition devices or the H\&E staining process. Nuclei segmentation is an important task, as it detects the nuclei cells over background tissue and gives rise to the topology, size, and count of nuclei which are determinant factors for cancer detection. Yet, it is a fairly time consuming task for pathologists, with reportedly high subjectivity. Computer Aided Diagnosis (CAD) tools empowered by modern Artificial Intelligence (AI) models enable the automation of nuclei segmentation. This can reduce the subjectivity in analysis and reading time. This paper provides an extensive review, beginning from earlier works use traditional image processing techniques and reaching up to modern approaches following the Deep Learning (DL) paradigm. Our review also focuses on the weak supervision aspect of the problem, motivated by the fact that annotated data is scarce. At the end, the advantages of different models and types of supervision are thoroughly discussed. Furthermore, we try to extrapolate and envision how future research lines will potentially be, so as to minimize the need for labeled data while maintaining high performance. Future methods should emphasize efficient and explainable models with a transparent underlying process so that physicians can trust their output.
Learning Scene Flow With Skeleton Guidance For 3D Action Recognition
Jun 23, 2023Abstract:Among the existing modalities for 3D action recognition, 3D flow has been poorly examined, although conveying rich motion information cues for human actions. Presumably, its susceptibility to noise renders it intractable, thus challenging the learning process within deep models. This work demonstrates the use of 3D flow sequence by a deep spatiotemporal model and further proposes an incremental two-level spatial attention mechanism, guided from skeleton domain, for emphasizing motion features close to the body joint areas and according to their informativeness. Towards this end, an extended deep skeleton model is also introduced to learn the most discriminant action motion dynamics, so as to estimate an informativeness score for each joint. Subsequently, a late fusion scheme is adopted between the two models for learning the high level cross-modal correlations. Experimental results on the currently largest and most challenging dataset NTU RGB+D, demonstrate the effectiveness of the proposed approach, achieving state-of-the-art results.
LGSQE: Lightweight Generated Sample Quality Evaluatoin
Nov 08, 2022



Abstract:Despite prolific work on evaluating generative models, little research has been done on the quality evaluation of an individual generated sample. To address this problem, a lightweight generated sample quality evaluation (LGSQE) method is proposed in this work. In the training stage of LGSQE, a binary classifier is trained on real and synthetic samples, where real and synthetic data are labeled by 0 and 1, respectively. In the inference stage, the classifier assigns soft labels (ranging from 0 to 1) to each generated sample. The value of soft label indicates the quality level; namely, the quality is better if its soft label is closer to 0. LGSQE can serve as a post-processing module for quality control. Furthermore, LGSQE can be used to evaluate the performance of generative models, such as accuracy, AUC, precision and recall, by aggregating sample-level quality. Experiments are conducted on CIFAR-10 and MNIST to demonstrate that LGSQE can preserve the same performance rank order as that predicted by the Frechet Inception Distance (FID) but with significantly lower complexity.
Statistical Attention Localization (SAL): Methodology and Application to Object Classification
Aug 03, 2022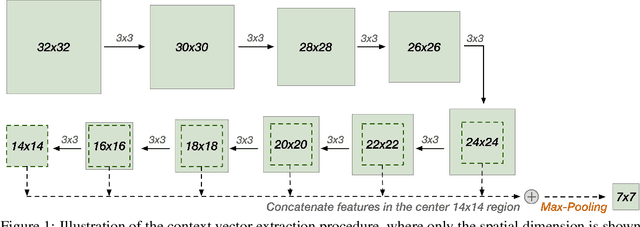
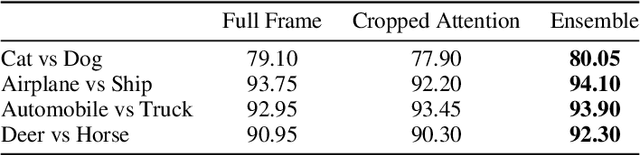
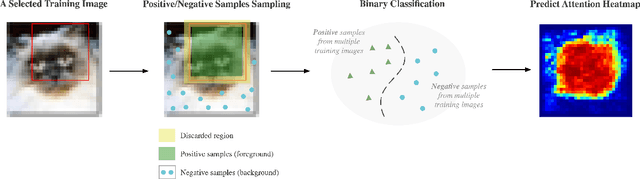
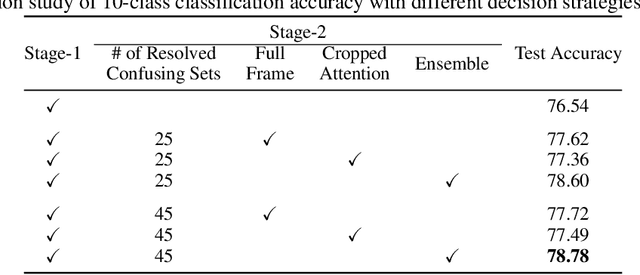
Abstract:A statistical attention localization (SAL) method is proposed to facilitate the object classification task in this work. SAL consists of three steps: 1) preliminary attention window selection via decision statistics, 2) attention map refinement, and 3) rectangular attention region finalization. SAL computes soft-decision scores of local squared windows and uses them to identify salient regions in Step 1. To accommodate object of various sizes and shapes, SAL refines the preliminary result and obtain an attention map of more flexible shape in Step 2. Finally, SAL yields a rectangular attention region using the refined attention map and bounding box regularization in Step 3. As an application, we adopt E-PixelHop, which is an object classification solution based on successive subspace learning (SSL), as the baseline. We apply SAL so as to obtain a cropped-out and resized attention region as an alternative input. Classification results of the whole image as well as the attention region are ensembled to achieve the highest classification accuracy. Experiments on the CIFAR-10 dataset are given to demonstrate the advantage of the SAL-assisted object classification method.
HUNIS: High-Performance Unsupervised Nuclei Instance Segmentation
Mar 28, 2022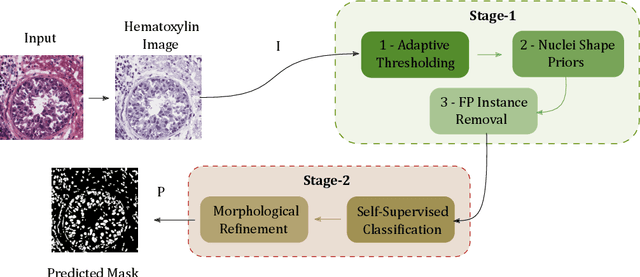
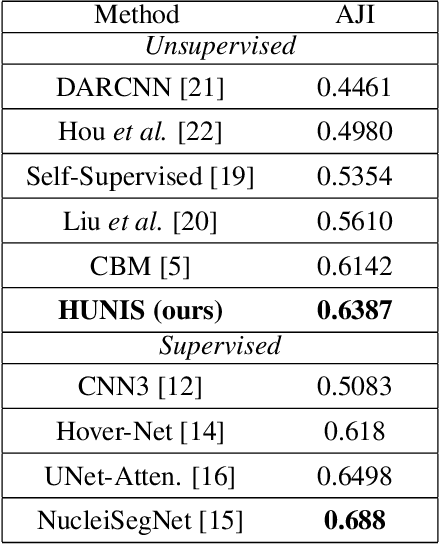
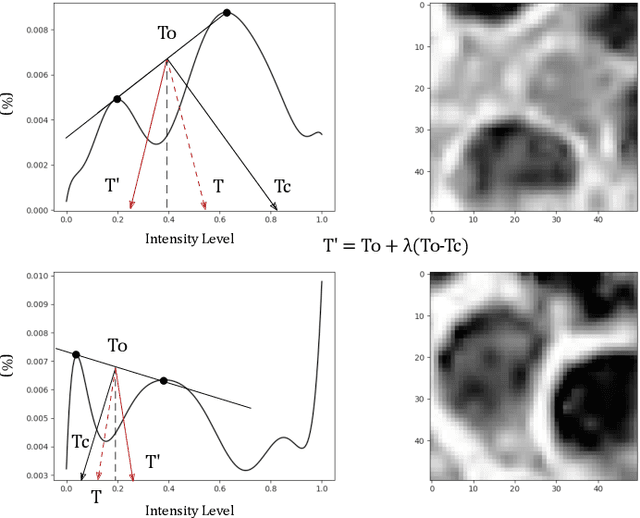
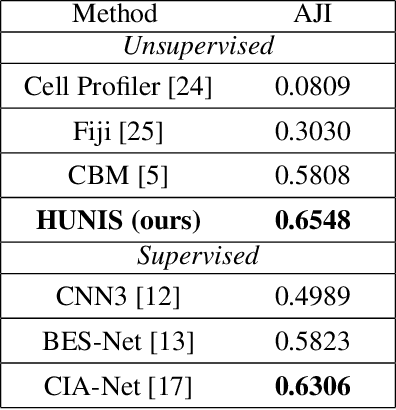
Abstract:A high-performance unsupervised nuclei instance segmentation (HUNIS) method is proposed in this work. HUNIS consists of two-stage block-wise operations. The first stage includes: 1) adaptive thresholding of pixel intensities, 2) incorporation of nuclei size/shape priors and 3) removal of false positive nuclei instances. Then, HUNIS conducts the second stage segmentation by receiving guidance from the first one. The second stage exploits the segmentation masks obtained in the first stage and leverages color and shape distributions for a more accurate segmentation. The main purpose of the two-stage design is to provide pixel-wise pseudo-labels from the first to the second stage. This self-supervision mechanism is novel and effective. Experimental results on the MoNuSeg dataset show that HUNIS outperforms all other unsupervised methods by a substantial margin. It also has a competitive standing among state-of-the-art supervised methods.
Unsupervised Data-Driven Nuclei Segmentation For Histology Images
Oct 14, 2021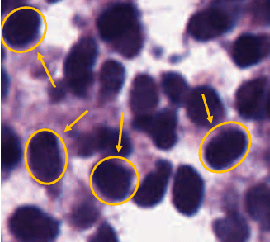
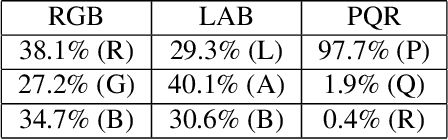
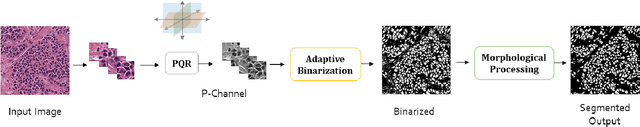
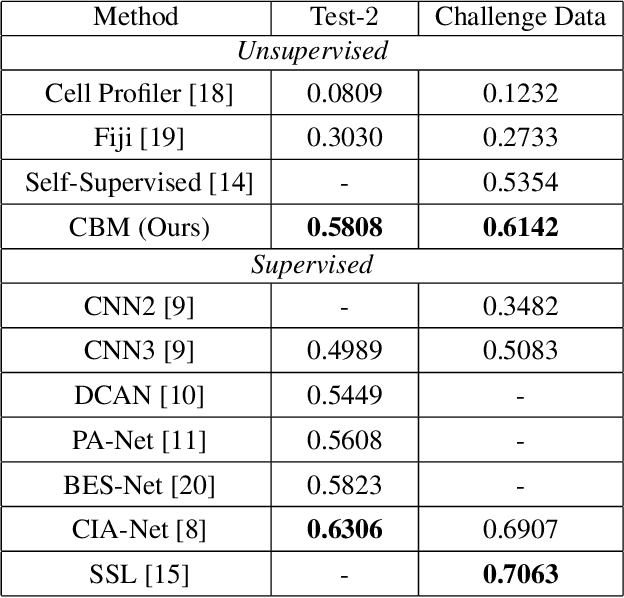
Abstract:An unsupervised data-driven nuclei segmentation method for histology images, called CBM, is proposed in this work. CBM consists of three modules applied in a block-wise manner: 1) data-driven color transform for energy compaction and dimension reduction, 2) data-driven binarization, and 3) incorporation of geometric priors with morphological processing. CBM comes from the first letter of the three modules - "Color transform", "Binarization" and "Morphological processing". Experiments on the MoNuSeg dataset validate the effectiveness of the proposed CBM method. CBM outperforms all other unsupervised methods and offers a competitive standing among supervised models based on the Aggregated Jaccard Index (AJI) metric.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge