Ting Cai
Constrained Best Arm Identification with Tests for Feasibility
Nov 12, 2025Abstract:Best arm identification (BAI) aims to identify the highest-performance arm among a set of $K$ arms by collecting stochastic samples from each arm. In real-world problems, the best arm needs to satisfy additional feasibility constraints. While there is limited prior work on BAI with feasibility constraints, they typically assume the performance and constraints are observed simultaneously on each pull of an arm. However, this assumption does not reflect most practical use cases, e.g., in drug discovery, we wish to find the most potent drug whose toxicity and solubility are below certain safety thresholds. These safety experiments can be conducted separately from the potency measurement. Thus, this requires designing BAI algorithms that not only decide which arm to pull but also decide whether to test for the arm's performance or feasibility. In this work, we study feasible BAI which allows a decision-maker to choose a tuple $(i,\ell)$, where $i\in [K]$ denotes an arm and $\ell$ denotes whether she wishes to test for its performance ($\ell=0$) or any of its $N$ feasibility constraints ($\ell\in[N]$). We focus on the fixed confidence setting, which is to identify the \textit{feasible} arm with the \textit{highest performance}, with a probability of at least $1-δ$. We propose an efficient algorithm and upper-bound its sample complexity, showing our algorithm can naturally adapt to the problem's difficulty and eliminate arms by worse performance or infeasibility, whichever is easier. We complement this upper bound with a lower bound showing that our algorithm is \textit{asymptotically ($δ\rightarrow 0$) optimal}. Finally, we empirically show that our algorithm outperforms other state-of-the-art BAI algorithms in both synthetic and real-world datasets.
RakutenAI-7B: Extending Large Language Models for Japanese
Mar 21, 2024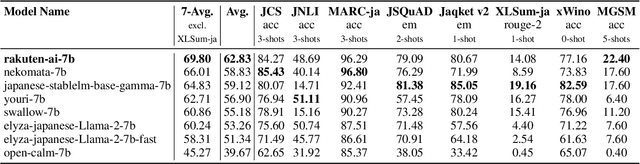
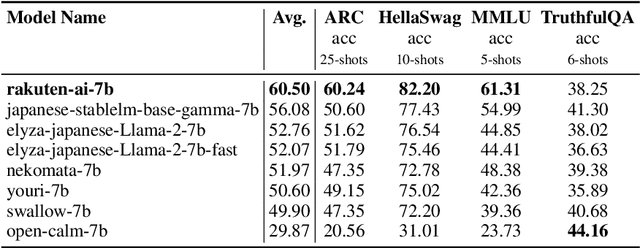
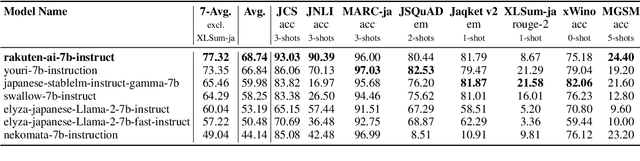
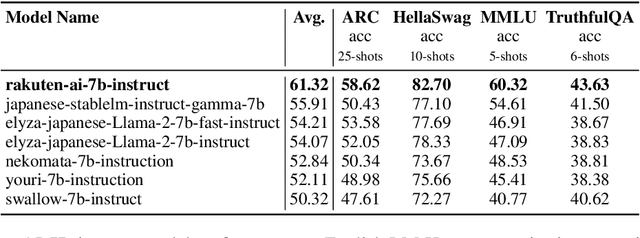
Abstract:We introduce RakutenAI-7B, a suite of Japanese-oriented large language models that achieve the best performance on the Japanese LM Harness benchmarks among the open 7B models. Along with the foundation model, we release instruction- and chat-tuned models, RakutenAI-7B-instruct and RakutenAI-7B-chat respectively, under the Apache 2.0 license.
Active Cost-aware Labeling of Streaming Data
Apr 13, 2023



Abstract:We study actively labeling streaming data, where an active learner is faced with a stream of data points and must carefully choose which of these points to label via an expensive experiment. Such problems frequently arise in applications such as healthcare and astronomy. We first study a setting when the data's inputs belong to one of $K$ discrete distributions and formalize this problem via a loss that captures the labeling cost and the prediction error. When the labeling cost is $B$, our algorithm, which chooses to label a point if the uncertainty is larger than a time and cost dependent threshold, achieves a worst-case upper bound of $O(B^{\frac{1}{3}} K^{\frac{1}{3}} T^{\frac{2}{3}})$ on the loss after $T$ rounds. We also provide a more nuanced upper bound which demonstrates that the algorithm can adapt to the arrival pattern, and achieves better performance when the arrival pattern is more favorable. We complement both upper bounds with matching lower bounds. We next study this problem when the inputs belong to a continuous domain and the output of the experiment is a smooth function with bounded RKHS norm. After $T$ rounds in $d$ dimensions, we show that the loss is bounded by $O(B^{\frac{1}{d+3}} T^{\frac{d+2}{d+3}})$ in an RKHS with a squared exponential kernel and by $O(B^{\frac{1}{2d+3}} T^{\frac{2d+2}{2d+3}})$ in an RKHS with a Mat\'ern kernel. Our empirical evaluation demonstrates that our method outperforms other baselines in several synthetic experiments and two real experiments in medicine and astronomy.
MVCNet: Multiview Contrastive Network for Unsupervised Representation Learning for 3D CT Lesions
Aug 18, 2021
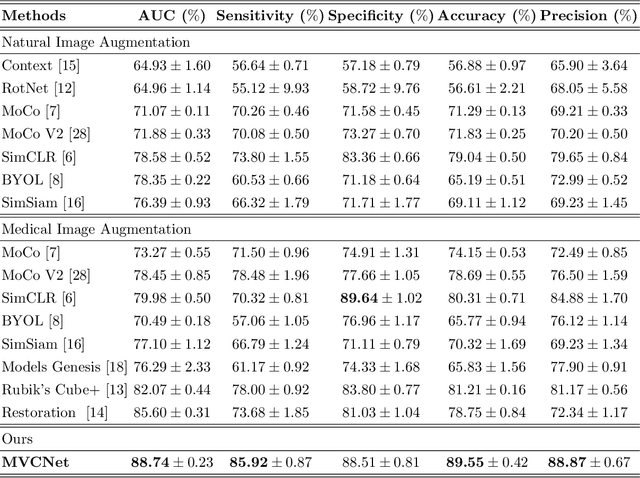
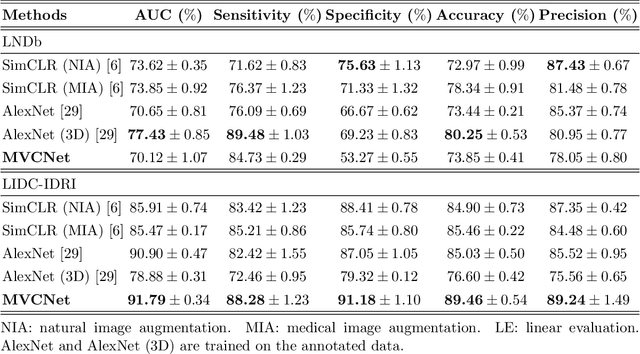
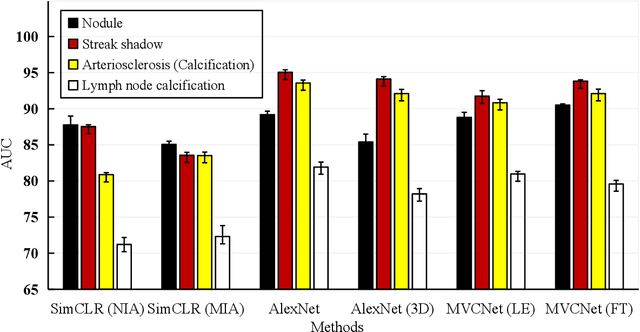
Abstract:\emph{Objective and Impact Statement}. With the renaissance of deep learning, automatic diagnostic systems for computed tomography (CT) have achieved many successful applications. However, they are mostly attributed to careful expert annotations, which are often scarce in practice. This drives our interest to the unsupervised representation learning. \emph{Introduction}. Recent studies have shown that self-supervised learning is an effective approach for learning representations, but most of them rely on the empirical design of transformations and pretext tasks. \emph{Methods}. To avoid the subjectivity associated with these methods, we propose the MVCNet, a novel unsupervised three dimensional (3D) representation learning method working in a transformation-free manner. We view each 3D lesion from different orientations to collect multiple two dimensional (2D) views. Then, an embedding function is learned by minimizing a contrastive loss so that the 2D views of the same 3D lesion are aggregated, and the 2D views of different lesions are separated. We evaluate the representations by training a simple classification head upon the embedding layer. \emph{Results}. Experimental results show that MVCNet achieves state-of-the-art accuracies on the LIDC-IDRI (89.55\%), LNDb (77.69\%) and TianChi (79.96\%) datasets for \emph{unsupervised representation learning}. When fine-tuned on 10\% of the labeled data, the accuracies are comparable to the supervised learning model (89.46\% vs. 85.03\%, 73.85\% vs. 73.44\%, 83.56\% vs. 83.34\% on the three datasets, respectively). \emph{Conclusion}. Results indicate the superiority of MVCNet in \emph{learning representations with limited annotations}.
Adversarially-Trained Nonnegative Matrix Factorization
Apr 10, 2021
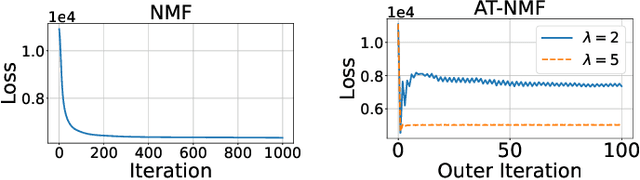


Abstract:We consider an adversarially-trained version of the nonnegative matrix factorization, a popular latent dimensionality reduction technique. In our formulation, an attacker adds an arbitrary matrix of bounded norm to the given data matrix. We design efficient algorithms inspired by adversarial training to optimize for dictionary and coefficient matrices with enhanced generalization abilities. Extensive simulations on synthetic and benchmark datasets demonstrate the superior predictive performance on matrix completion tasks of our proposed method compared to state-of-the-art competitors, including other variants of adversarial nonnegative matrix factorization.
Investigating Critical Risk Factors in Liver Cancer Prediction
Feb 03, 2021


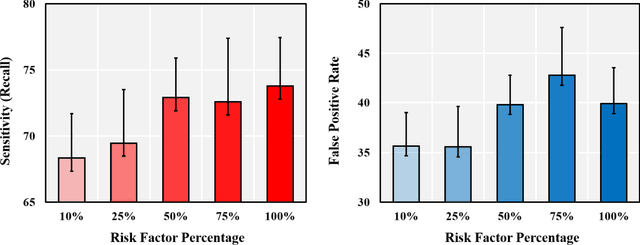
Abstract:We exploit liver cancer prediction model using machine learning algorithms based on epidemiological data of over 55 thousand peoples from 2014 to the present. The best performance is an AUC of 0.71. We analyzed model parameters to investigate critical risk factors that contribute the most to prediction.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge