Penghua Zhai
Spiral Contrastive Learning: An Efficient 3D Representation Learning Method for Unannotated CT Lesions
Aug 23, 2022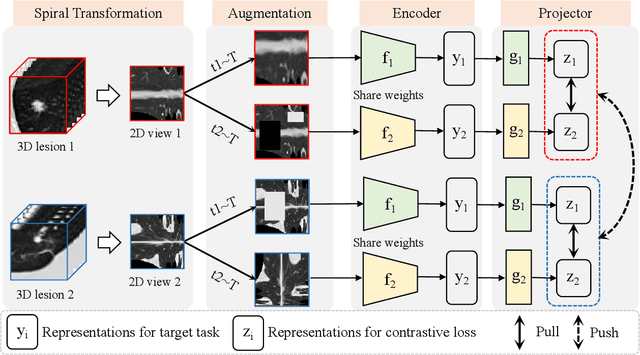
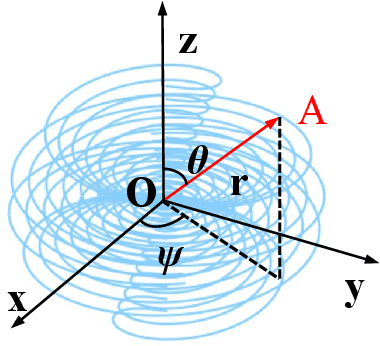
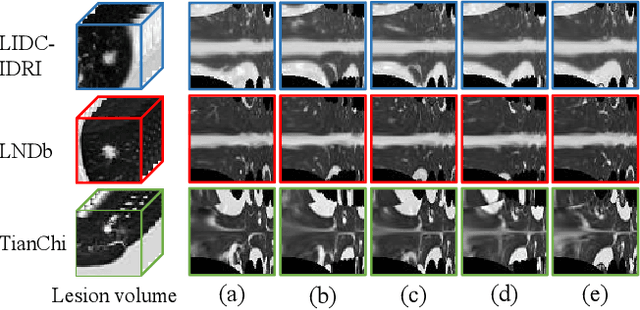
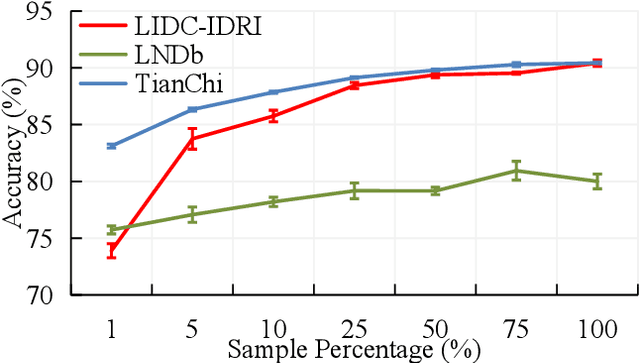
Abstract:Computed tomography (CT) samples with pathological annotations are difficult to obtain. As a result, the computer-aided diagnosis (CAD) algorithms are trained on small datasets (e.g., LIDC-IDRI with 1,018 samples), limiting their accuracies and reliability. In the past five years, several works have tailored for unsupervised representations of CT lesions via two-dimensional (2D) and three-dimensional (3D) self-supervised learning (SSL) algorithms. The 2D algorithms have difficulty capturing 3D information, and existing 3D algorithms are computationally heavy. Light-weight 3D SSL remains the boundary to explore. In this paper, we propose the spiral contrastive learning (SCL), which yields 3D representations in a computationally efficient manner. SCL first transforms 3D lesions to the 2D plane using an information-preserving spiral transformation, and then learn transformation-invariant features using 2D contrastive learning. For the augmentation, we consider natural image augmentations and medical image augmentations. We evaluate SCL by training a classification head upon the embedding layer. Experimental results show that SCL achieves state-of-the-art accuracy on LIDC-IDRI (89.72%), LNDb (82.09%) and TianChi (90.16%) for unsupervised representation learning. With 10% annotated data for fine-tune, the performance of SCL is comparable to that of supervised learning algorithms (85.75% vs. 85.03% on LIDC-IDRI, 78.20% vs. 73.44% on LNDb and 87.85% vs. 83.34% on TianChi, respectively). Meanwhile, SCL reduces the computational effort by 66.98% compared to other 3D SSL algorithms, demonstrating the efficiency of the proposed method in unsupervised pre-training.
MVCNet: Multiview Contrastive Network for Unsupervised Representation Learning for 3D CT Lesions
Aug 18, 2021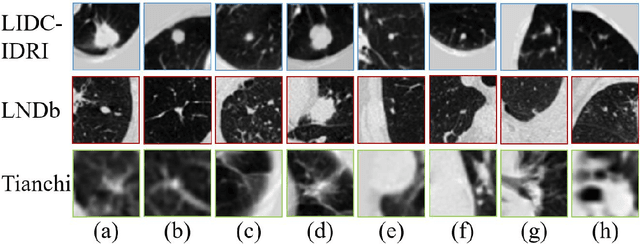
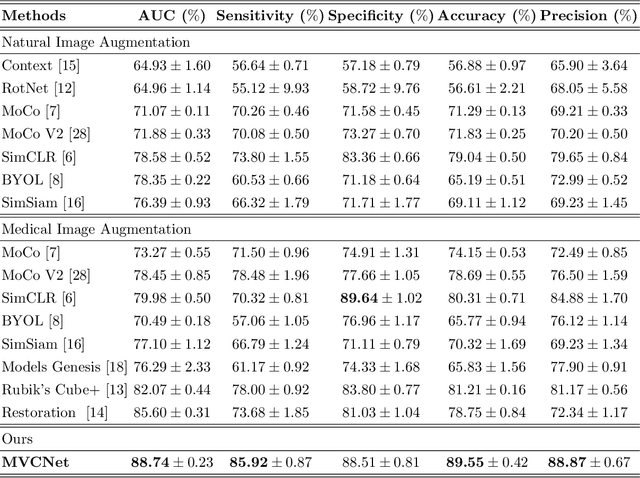
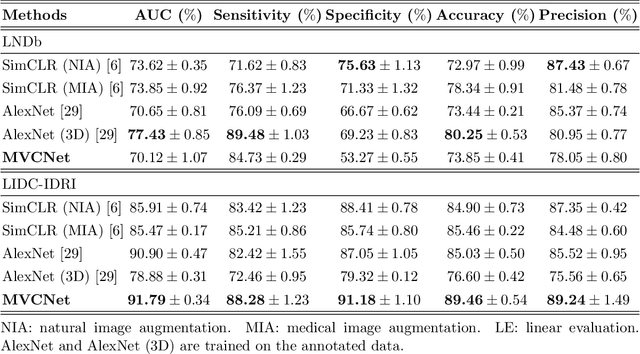
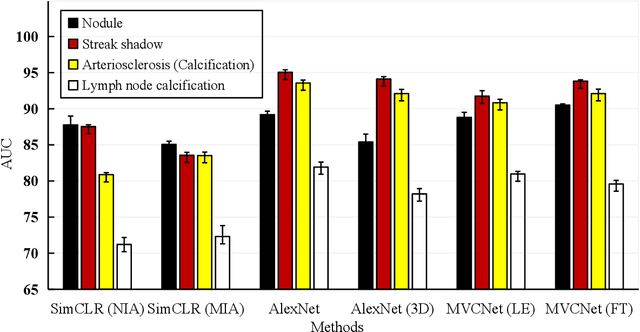
Abstract:\emph{Objective and Impact Statement}. With the renaissance of deep learning, automatic diagnostic systems for computed tomography (CT) have achieved many successful applications. However, they are mostly attributed to careful expert annotations, which are often scarce in practice. This drives our interest to the unsupervised representation learning. \emph{Introduction}. Recent studies have shown that self-supervised learning is an effective approach for learning representations, but most of them rely on the empirical design of transformations and pretext tasks. \emph{Methods}. To avoid the subjectivity associated with these methods, we propose the MVCNet, a novel unsupervised three dimensional (3D) representation learning method working in a transformation-free manner. We view each 3D lesion from different orientations to collect multiple two dimensional (2D) views. Then, an embedding function is learned by minimizing a contrastive loss so that the 2D views of the same 3D lesion are aggregated, and the 2D views of different lesions are separated. We evaluate the representations by training a simple classification head upon the embedding layer. \emph{Results}. Experimental results show that MVCNet achieves state-of-the-art accuracies on the LIDC-IDRI (89.55\%), LNDb (77.69\%) and TianChi (79.96\%) datasets for \emph{unsupervised representation learning}. When fine-tuned on 10\% of the labeled data, the accuracies are comparable to the supervised learning model (89.46\% vs. 85.03\%, 73.85\% vs. 73.44\%, 83.56\% vs. 83.34\% on the three datasets, respectively). \emph{Conclusion}. Results indicate the superiority of MVCNet in \emph{learning representations with limited annotations}.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge