Huaiwei Cong
Faint Features Tell: Automatic Vertebrae Fracture Screening Assisted by Contrastive Learning
Aug 23, 2022



Abstract:Long-term vertebral fractures severely affect the life quality of patients, causing kyphotic, lumbar deformity and even paralysis. Computed tomography (CT) is a common clinical examination to screen for this disease at early stages. However, the faint radiological appearances and unspecific symptoms lead to a high risk of missed diagnosis. In particular, the mild fractures and normal controls are quite difficult to distinguish for deep learning models and inexperienced doctors. In this paper, we argue that reinforcing the faint fracture features to encourage the inter-class separability is the key to improving the accuracy. Motivated by this, we propose a supervised contrastive learning based model to estimate Genent's Grade of vertebral fracture with CT scans. The supervised contrastive learning, as an auxiliary task, narrows the distance of features within the same class while pushing others away, which enhances the model's capability of capturing subtle features of vertebral fractures. Considering the lack of datasets in this field, we construct a database including 208 samples annotated by experienced radiologists. Our method has a specificity of 99\% and a sensitivity of 85\% in binary classification, and a macio-F1 of 77\% in multi-classification, indicating that contrastive learning significantly improves the accuracy of vertebrae fracture screening, especially for the mild fractures and normal controls. Our desensitized data and codes will be made publicly available for the community.
Structure Regularized Attentive Network for Automatic Femoral Head Necrosis Diagnosis and Localization
Aug 23, 2022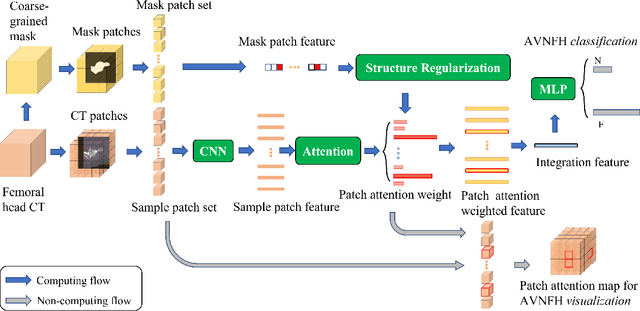
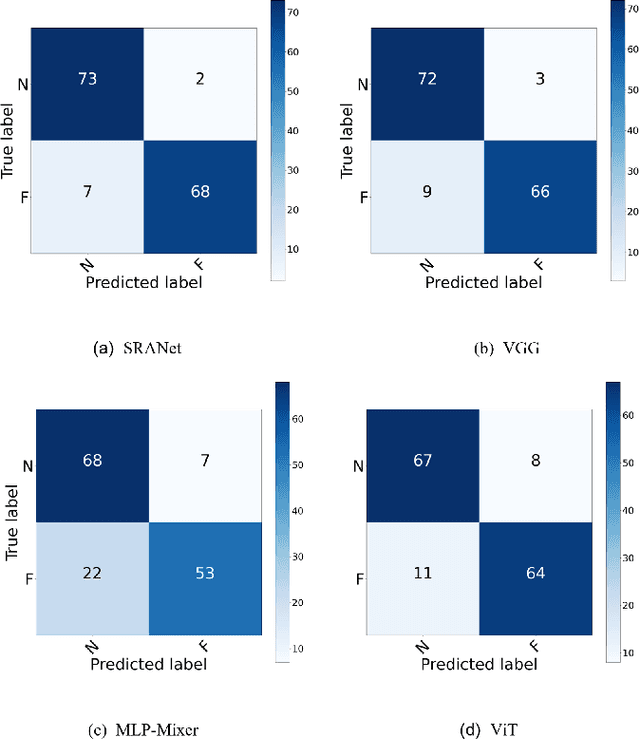
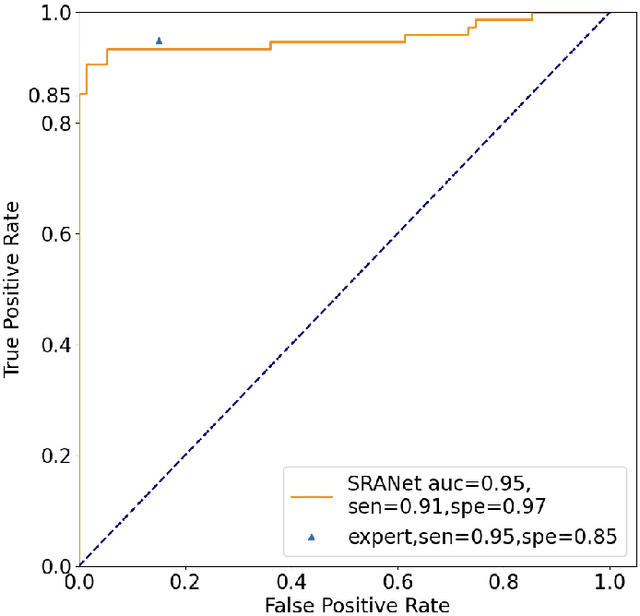
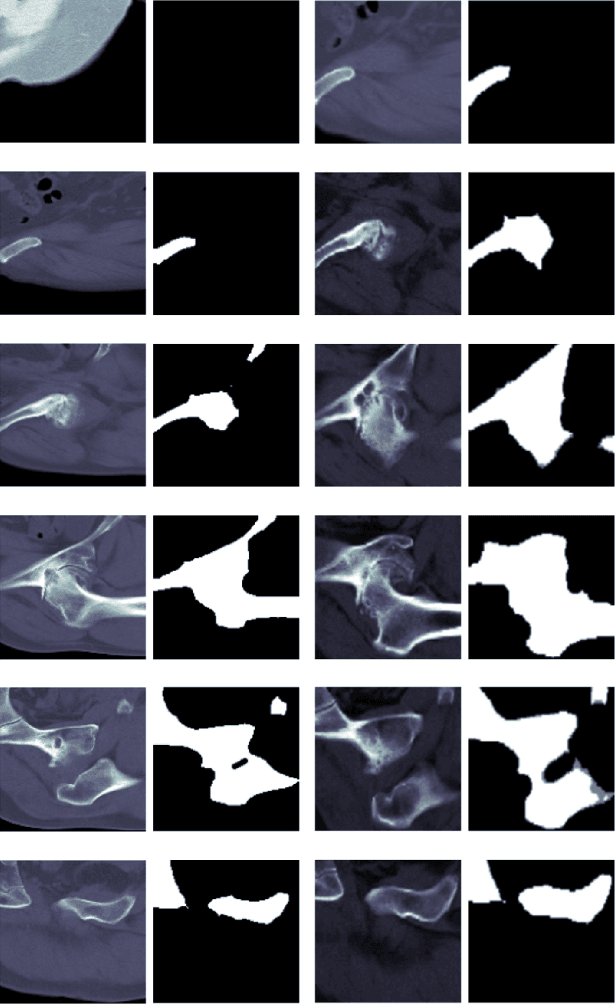
Abstract:In recent years, several works have adopted the convolutional neural network (CNN) to diagnose the avascular necrosis of the femoral head (AVNFH) based on X-ray images or magnetic resonance imaging (MRI). However, due to the tissue overlap, X-ray images are difficult to provide fine-grained features for early diagnosis. MRI, on the other hand, has a long imaging time, is more expensive, making it impractical in mass screening. Computed tomography (CT) shows layer-wise tissues, is faster to image, and is less costly than MRI. However, to our knowledge, there is no work on CT-based automated diagnosis of AVNFH. In this work, we collected and labeled a large-scale dataset for AVNFH ranking. In addition, existing end-to-end CNNs only yields the classification result and are difficult to provide more information for doctors in diagnosis. To address this issue, we propose the structure regularized attentive network (SRANet), which is able to highlight the necrotic regions during classification based on patch attention. SRANet extracts features in chunks of images, obtains weight via the attention mechanism to aggregate the features, and constrains them by a structural regularizer with prior knowledge to improve the generalization. SRANet was evaluated on our AVNFH-CT dataset. Experimental results show that SRANet is superior to CNNs for AVNFH classification, moreover, it can localize lesions and provide more information to assist doctors in diagnosis. Our codes are made public at https://github.com/tomas-lilingfeng/SRANet.
MVCNet: Multiview Contrastive Network for Unsupervised Representation Learning for 3D CT Lesions
Aug 18, 2021
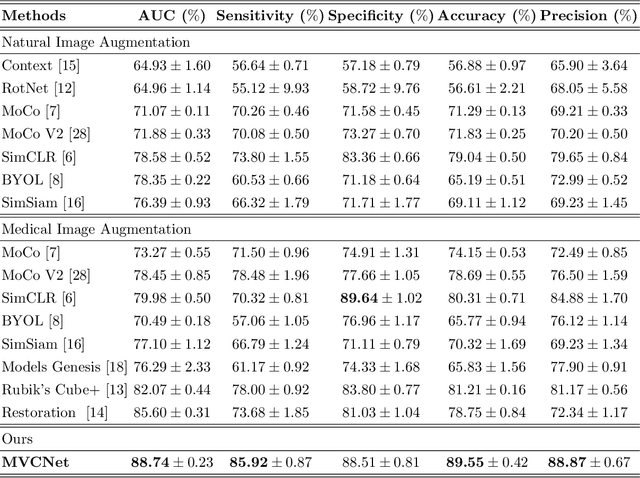
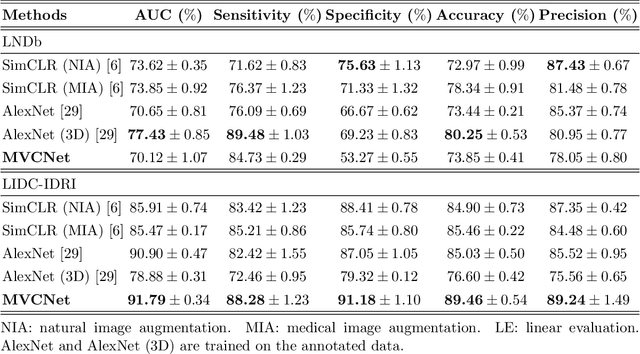
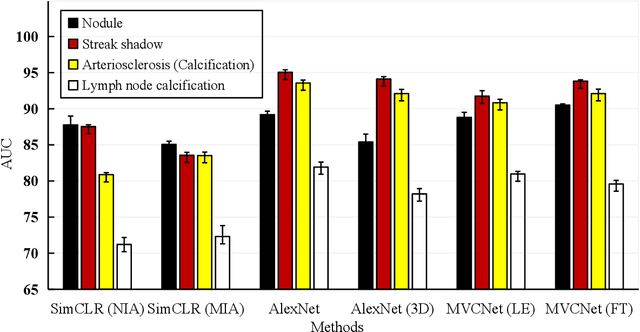
Abstract:\emph{Objective and Impact Statement}. With the renaissance of deep learning, automatic diagnostic systems for computed tomography (CT) have achieved many successful applications. However, they are mostly attributed to careful expert annotations, which are often scarce in practice. This drives our interest to the unsupervised representation learning. \emph{Introduction}. Recent studies have shown that self-supervised learning is an effective approach for learning representations, but most of them rely on the empirical design of transformations and pretext tasks. \emph{Methods}. To avoid the subjectivity associated with these methods, we propose the MVCNet, a novel unsupervised three dimensional (3D) representation learning method working in a transformation-free manner. We view each 3D lesion from different orientations to collect multiple two dimensional (2D) views. Then, an embedding function is learned by minimizing a contrastive loss so that the 2D views of the same 3D lesion are aggregated, and the 2D views of different lesions are separated. We evaluate the representations by training a simple classification head upon the embedding layer. \emph{Results}. Experimental results show that MVCNet achieves state-of-the-art accuracies on the LIDC-IDRI (89.55\%), LNDb (77.69\%) and TianChi (79.96\%) datasets for \emph{unsupervised representation learning}. When fine-tuned on 10\% of the labeled data, the accuracies are comparable to the supervised learning model (89.46\% vs. 85.03\%, 73.85\% vs. 73.44\%, 83.56\% vs. 83.34\% on the three datasets, respectively). \emph{Conclusion}. Results indicate the superiority of MVCNet in \emph{learning representations with limited annotations}.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge