Enwei Zhu
Recognizing Nested Entities from Flat Supervision: A New NER Subtask, Feasibility and Challenges
Nov 01, 2022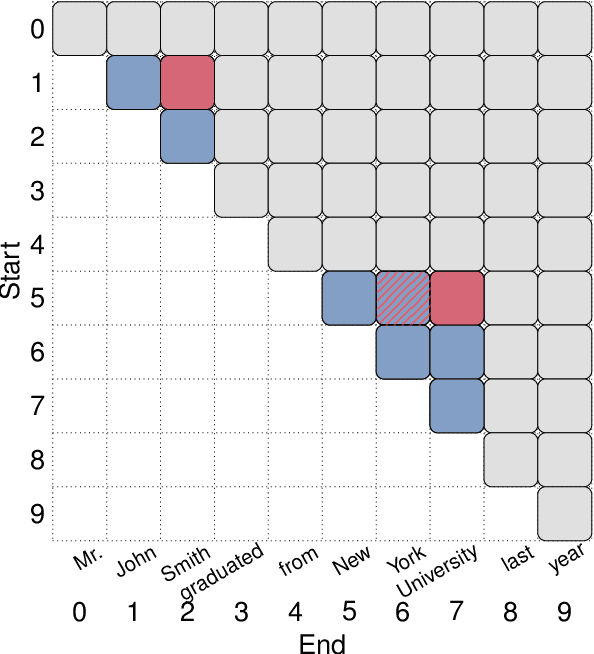
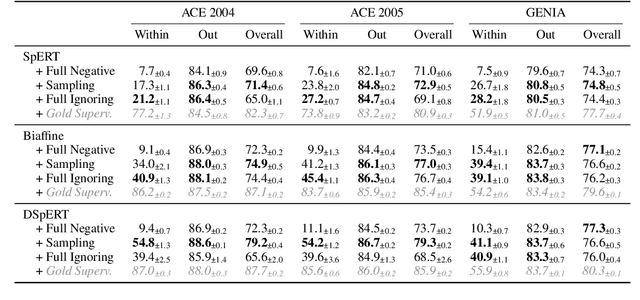
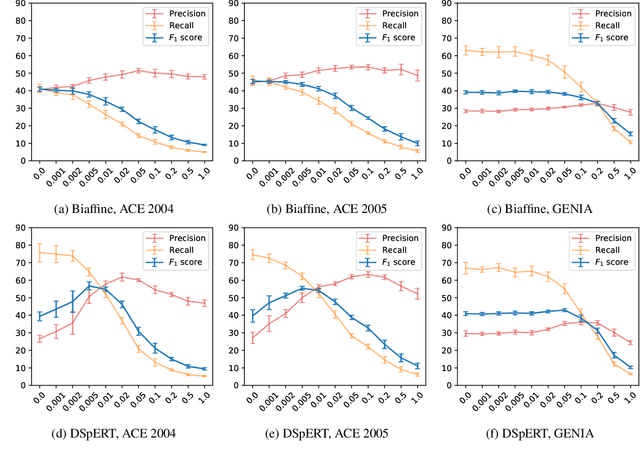
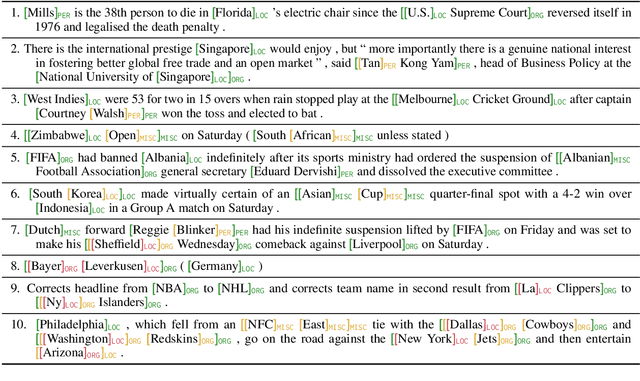
Abstract:Many recent named entity recognition (NER) studies criticize flat NER for its non-overlapping assumption, and switch to investigating nested NER. However, existing nested NER models heavily rely on training data annotated with nested entities, while labeling such data is costly. This study proposes a new subtask, nested-from-flat NER, which corresponds to a realistic application scenario: given data annotated with flat entities only, one may still desire the trained model capable of recognizing nested entities. To address this task, we train span-based models and deliberately ignore the spans nested inside labeled entities, since these spans are possibly unlabeled entities. With nested entities removed from the training data, our model achieves 54.8%, 54.2% and 41.1% F1 scores on the subset of spans within entities on ACE 2004, ACE 2005 and GENIA, respectively. This suggests the effectiveness of our approach and the feasibility of the task. In addition, the model's performance on flat entities is entirely unaffected. We further manually annotate the nested entities in the test set of CoNLL 2003, creating a nested-from-flat NER benchmark. Analysis results show that the main challenges stem from the data and annotation inconsistencies between the flat and nested entities.
Deep Span Representations for Named Entity Recognition
Oct 09, 2022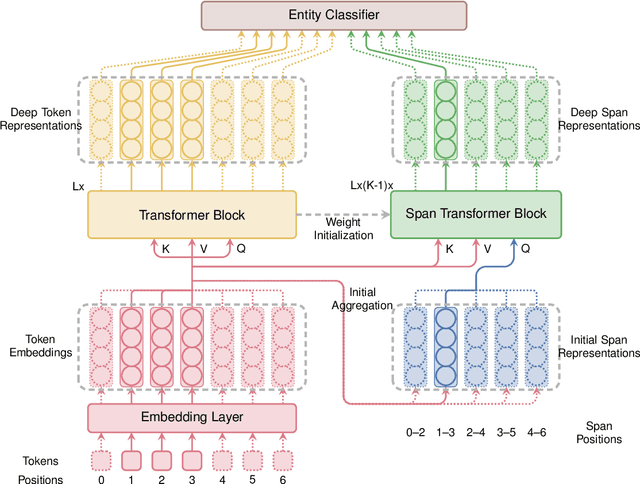
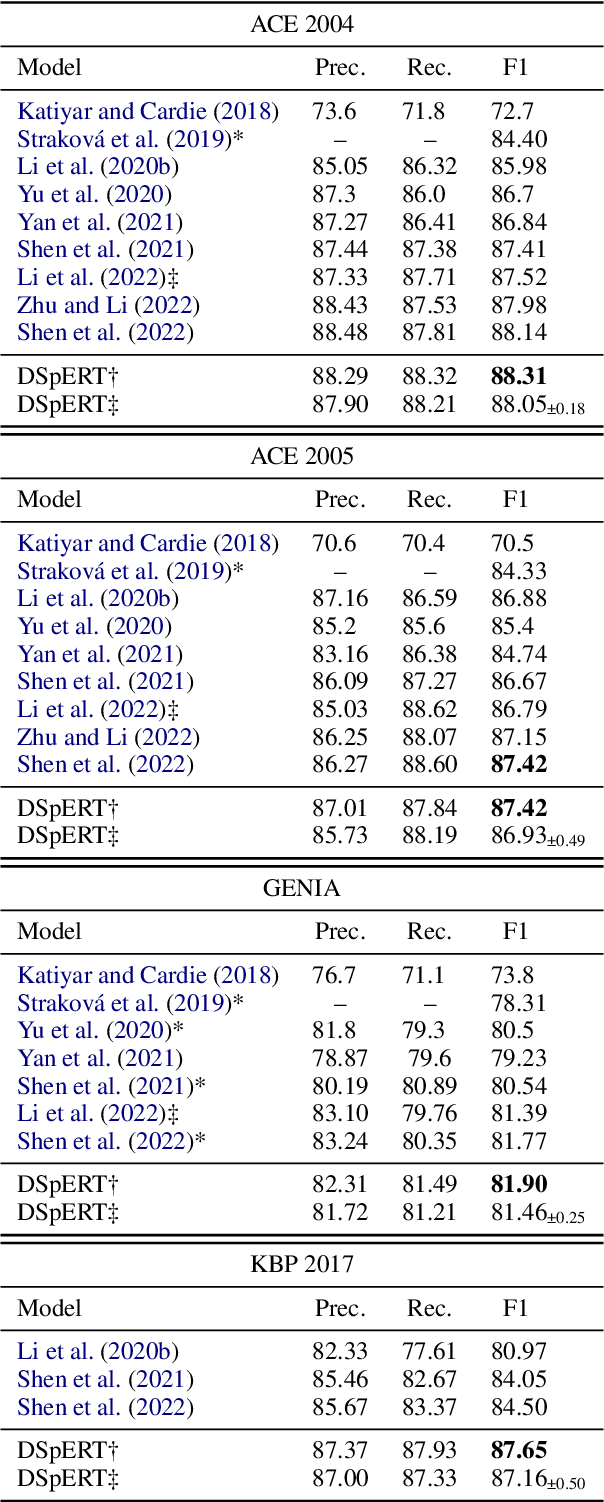

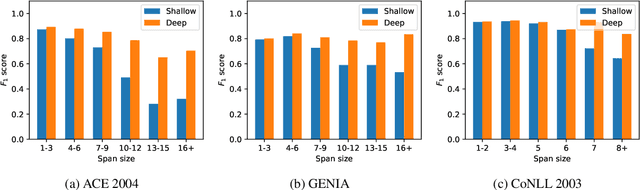
Abstract:Span-based models are one of the most straightforward methods for named entity recognition (NER). Existing span-based NER systems shallowly aggregate the token representations to span representations. However, this typically results in significant ineffectiveness for long-span entities, a coupling between the representations of overlapping spans, and ultimately a performance degradation. In this study, we propose DSpERT (Deep Span Encoder Representations from Transformers), which comprises a standard Transformer and a span Transformer. The latter uses low-layered span representations as queries, and aggregates the token representations as keys and values, layer by layer from bottom to top. Thus, DSpERT produces span representations of deep semantics. With weight initialization from pretrained language models, DSpERT achieves performance higher than or competitive with recent state-of-the-art systems on eight NER benchmarks. Experimental results verify the importance of the depth for span representations, and show that DSpERT performs particularly well on long-span entities and nested structures. Further, the deep span representations are well structured and easily separable in the feature space.
Spiral Contrastive Learning: An Efficient 3D Representation Learning Method for Unannotated CT Lesions
Aug 23, 2022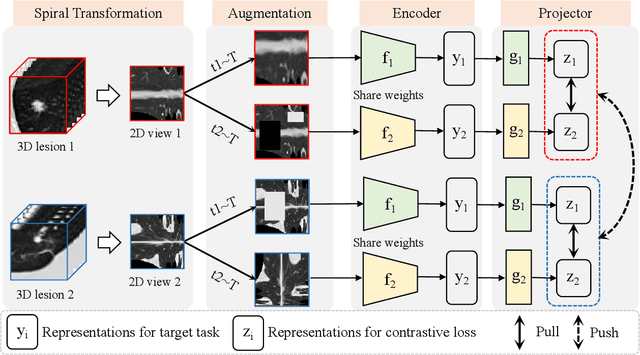
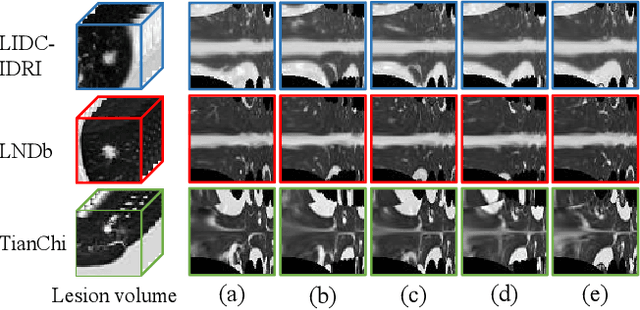
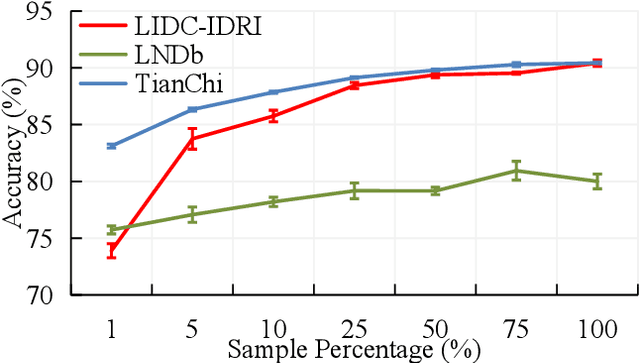
Abstract:Computed tomography (CT) samples with pathological annotations are difficult to obtain. As a result, the computer-aided diagnosis (CAD) algorithms are trained on small datasets (e.g., LIDC-IDRI with 1,018 samples), limiting their accuracies and reliability. In the past five years, several works have tailored for unsupervised representations of CT lesions via two-dimensional (2D) and three-dimensional (3D) self-supervised learning (SSL) algorithms. The 2D algorithms have difficulty capturing 3D information, and existing 3D algorithms are computationally heavy. Light-weight 3D SSL remains the boundary to explore. In this paper, we propose the spiral contrastive learning (SCL), which yields 3D representations in a computationally efficient manner. SCL first transforms 3D lesions to the 2D plane using an information-preserving spiral transformation, and then learn transformation-invariant features using 2D contrastive learning. For the augmentation, we consider natural image augmentations and medical image augmentations. We evaluate SCL by training a classification head upon the embedding layer. Experimental results show that SCL achieves state-of-the-art accuracy on LIDC-IDRI (89.72%), LNDb (82.09%) and TianChi (90.16%) for unsupervised representation learning. With 10% annotated data for fine-tune, the performance of SCL is comparable to that of supervised learning algorithms (85.75% vs. 85.03% on LIDC-IDRI, 78.20% vs. 73.44% on LNDb and 87.85% vs. 83.34% on TianChi, respectively). Meanwhile, SCL reduces the computational effort by 66.98% compared to other 3D SSL algorithms, demonstrating the efficiency of the proposed method in unsupervised pre-training.
Boundary Smoothing for Named Entity Recognition
Apr 26, 2022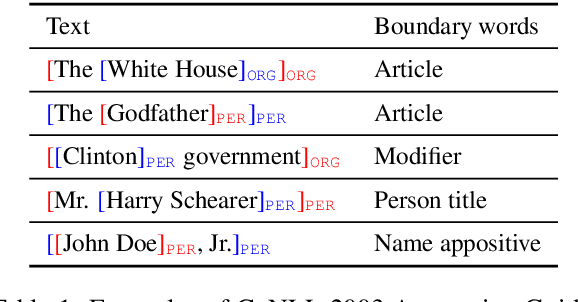
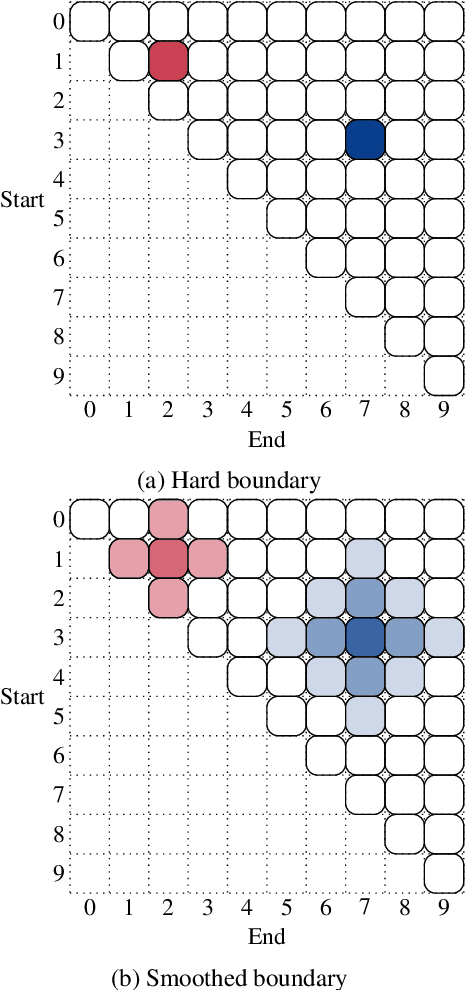
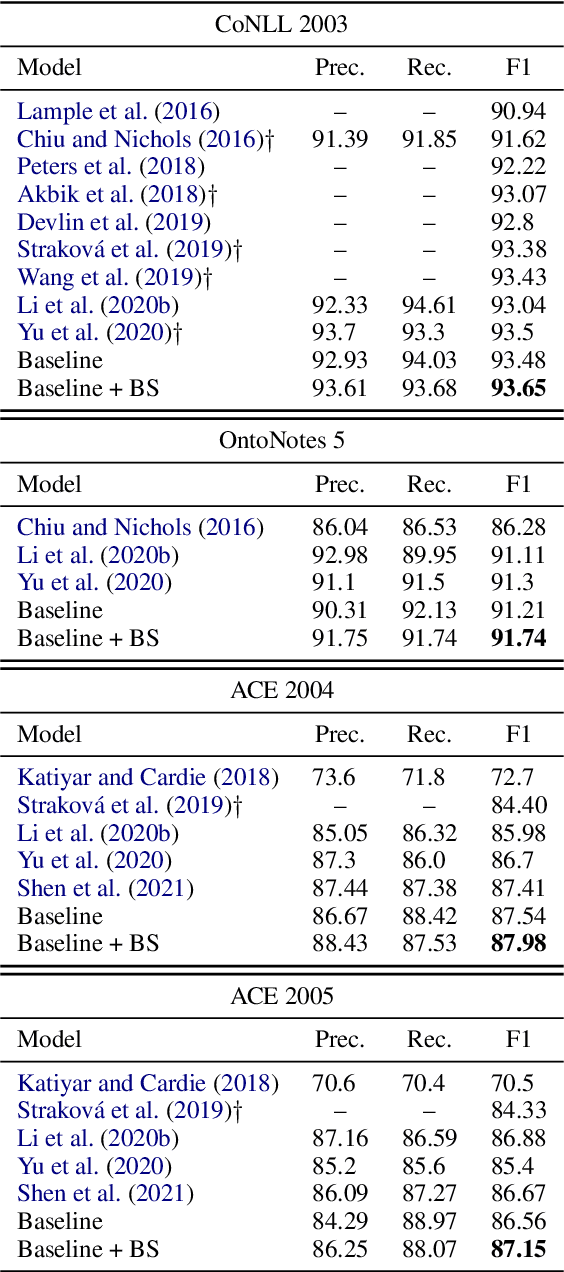
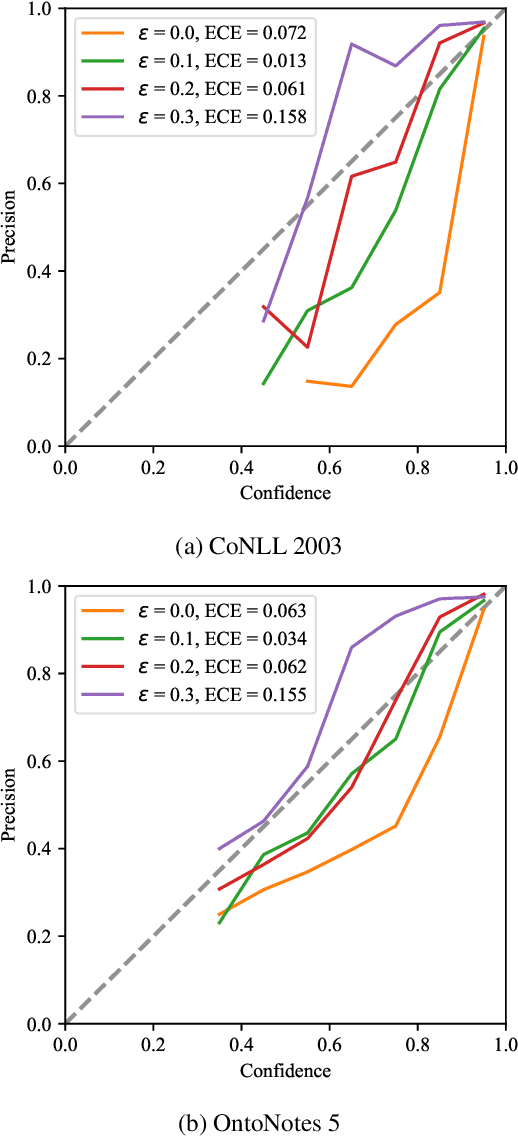
Abstract:Neural named entity recognition (NER) models may easily encounter the over-confidence issue, which degrades the performance and calibration. Inspired by label smoothing and driven by the ambiguity of boundary annotation in NER engineering, we propose boundary smoothing as a regularization technique for span-based neural NER models. It re-assigns entity probabilities from annotated spans to the surrounding ones. Built on a simple but strong baseline, our model achieves results better than or competitive with previous state-of-the-art systems on eight well-known NER benchmarks. Further empirical analysis suggests that boundary smoothing effectively mitigates over-confidence, improves model calibration, and brings flatter neural minima and more smoothed loss landscapes.
A Unified Framework of Medical Information Annotation and Extraction for Chinese Clinical Text
Mar 08, 2022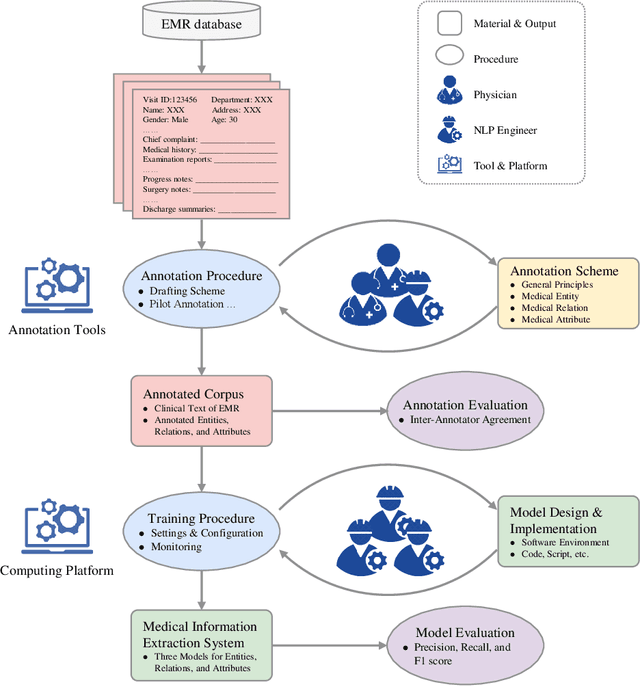
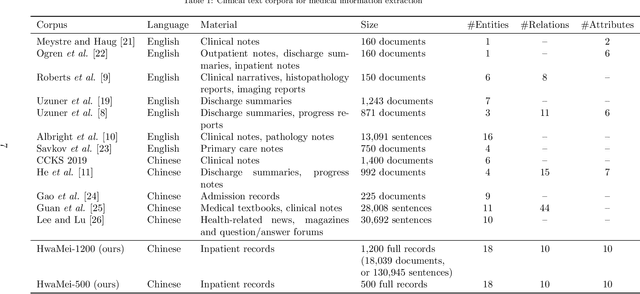
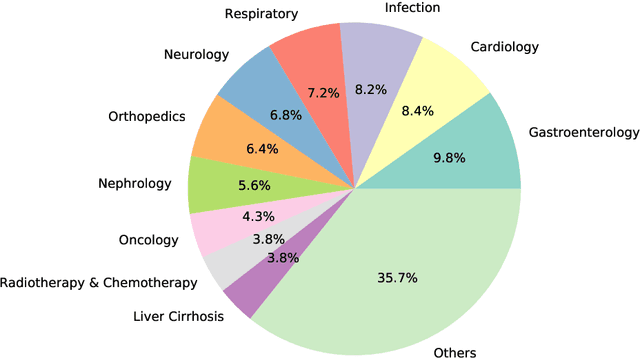
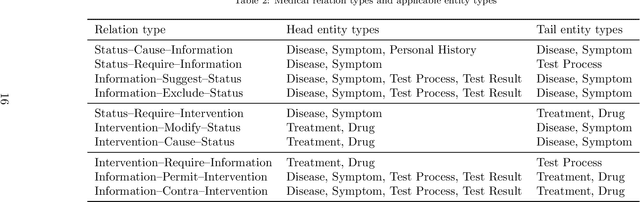
Abstract:Medical information extraction consists of a group of natural language processing (NLP) tasks, which collaboratively convert clinical text to pre-defined structured formats. Current state-of-the-art (SOTA) NLP models are highly integrated with deep learning techniques and thus require massive annotated linguistic data. This study presents an engineering framework of medical entity recognition, relation extraction and attribute extraction, which are unified in annotation, modeling and evaluation. Specifically, the annotation scheme is comprehensive, and compatible between tasks, especially for the medical relations. The resulted annotated corpus includes 1,200 full medical records (or 18,039 broken-down documents), and achieves inter-annotator agreements (IAAs) of 94.53%, 73.73% and 91.98% F 1 scores for the three tasks. Three task-specific neural network models are developed within a shared structure, and enhanced by SOTA NLP techniques, i.e., pre-trained language models. Experimental results show that the system can retrieve medical entities, relations and attributes with F 1 scores of 93.47%, 67.14% and 90.89%, respectively. This study, in addition to our publicly released annotation scheme and code, provides solid and practical engineering experience of developing an integrated medical information extraction system.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge