Baolian Qi
Deep 3D Vessel Segmentation based on Cross Transformer Network
Aug 23, 2022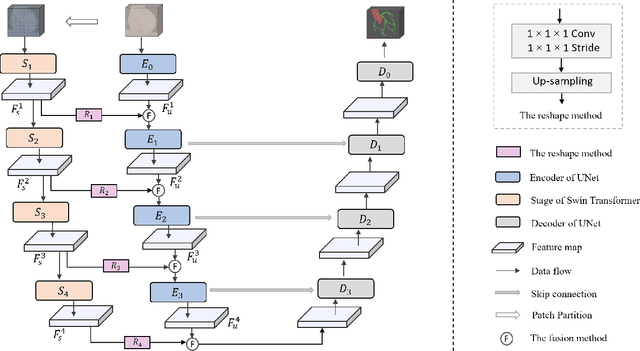
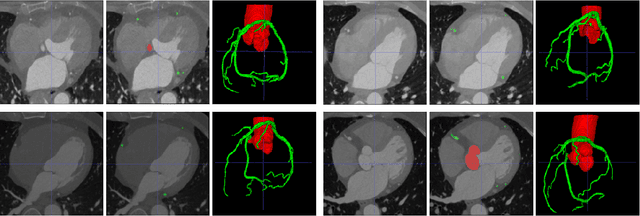
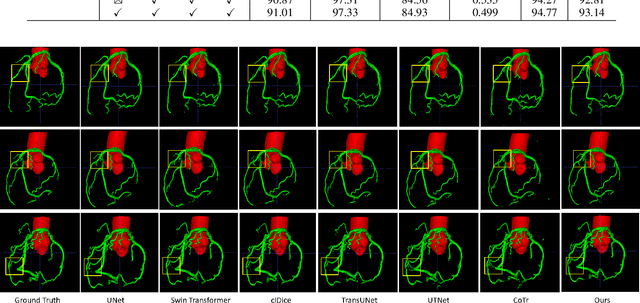
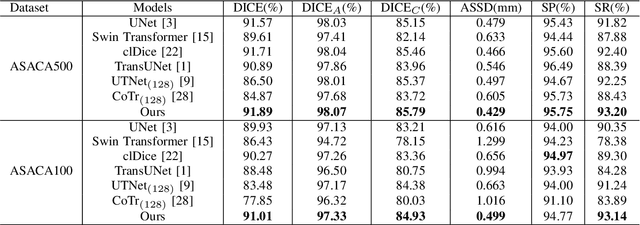
Abstract:The coronary microvascular disease poses a great threat to human health. Computer-aided analysis/diagnosis systems help physicians intervene in the disease at early stages, where 3D vessel segmentation is a fundamental step. However, there is a lack of carefully annotated dataset to support algorithm development and evaluation. On the other hand, the commonly-used U-Net structures often yield disconnected and inaccurate segmentation results, especially for small vessel structures. In this paper, motivated by the data scarcity, we first construct two large-scale vessel segmentation datasets consisting of 100 and 500 computed tomography (CT) volumes with pixel-level annotations by experienced radiologists. To enhance the U-Net, we further propose the cross transformer network (CTN) for fine-grained vessel segmentation. In CTN, a transformer module is constructed in parallel to a U-Net to learn long-distance dependencies between different anatomical regions; and these dependencies are communicated to the U-Net at multiple stages to endow it with global awareness. Experimental results on the two in-house datasets indicate that this hybrid model alleviates unexpected disconnections by considering topological information across regions. Our codes, together with the trained models are made publicly available at https://github.com/qibaolian/ctn.
Spiral Contrastive Learning: An Efficient 3D Representation Learning Method for Unannotated CT Lesions
Aug 23, 2022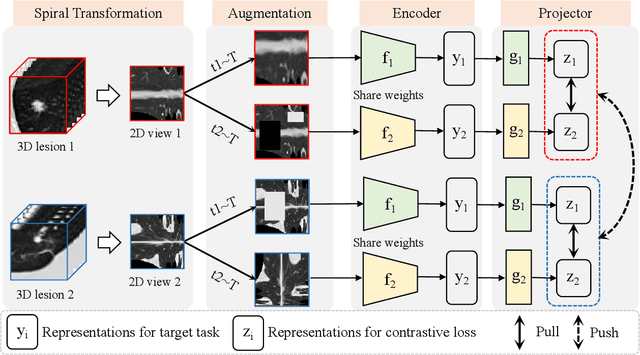
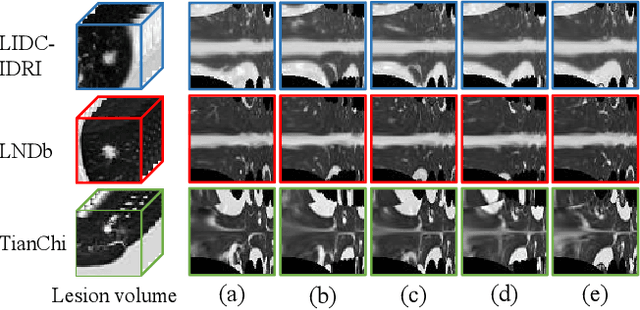
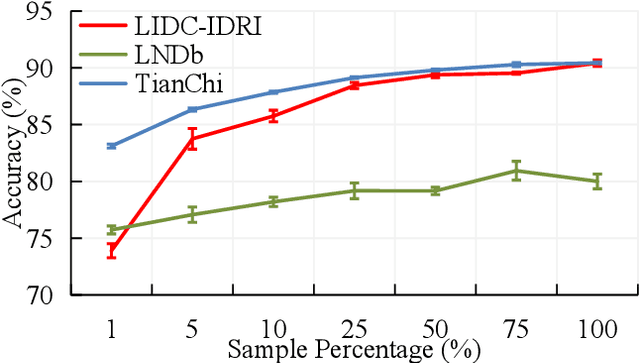
Abstract:Computed tomography (CT) samples with pathological annotations are difficult to obtain. As a result, the computer-aided diagnosis (CAD) algorithms are trained on small datasets (e.g., LIDC-IDRI with 1,018 samples), limiting their accuracies and reliability. In the past five years, several works have tailored for unsupervised representations of CT lesions via two-dimensional (2D) and three-dimensional (3D) self-supervised learning (SSL) algorithms. The 2D algorithms have difficulty capturing 3D information, and existing 3D algorithms are computationally heavy. Light-weight 3D SSL remains the boundary to explore. In this paper, we propose the spiral contrastive learning (SCL), which yields 3D representations in a computationally efficient manner. SCL first transforms 3D lesions to the 2D plane using an information-preserving spiral transformation, and then learn transformation-invariant features using 2D contrastive learning. For the augmentation, we consider natural image augmentations and medical image augmentations. We evaluate SCL by training a classification head upon the embedding layer. Experimental results show that SCL achieves state-of-the-art accuracy on LIDC-IDRI (89.72%), LNDb (82.09%) and TianChi (90.16%) for unsupervised representation learning. With 10% annotated data for fine-tune, the performance of SCL is comparable to that of supervised learning algorithms (85.75% vs. 85.03% on LIDC-IDRI, 78.20% vs. 73.44% on LNDb and 87.85% vs. 83.34% on TianChi, respectively). Meanwhile, SCL reduces the computational effort by 66.98% compared to other 3D SSL algorithms, demonstrating the efficiency of the proposed method in unsupervised pre-training.
Computer-aided Tuberculosis Diagnosis with Attribute Reasoning Assistance
Jul 01, 2022



Abstract:Although deep learning algorithms have been intensively developed for computer-aided tuberculosis diagnosis (CTD), they mainly depend on carefully annotated datasets, leading to much time and resource consumption. Weakly supervised learning (WSL), which leverages coarse-grained labels to accomplish fine-grained tasks, has the potential to solve this problem. In this paper, we first propose a new large-scale tuberculosis (TB) chest X-ray dataset, namely the tuberculosis chest X-ray attribute dataset (TBX-Att), and then establish an attribute-assisted weakly-supervised framework to classify and localize TB by leveraging the attribute information to overcome the insufficiency of supervision in WSL scenarios. Specifically, first, the TBX-Att dataset contains 2000 X-ray images with seven kinds of attributes for TB relational reasoning, which are annotated by experienced radiologists. It also includes the public TBX11K dataset with 11200 X-ray images to facilitate weakly supervised detection. Second, we exploit a multi-scale feature interaction model for TB area classification and detection with attribute relational reasoning. The proposed model is evaluated on the TBX-Att dataset and will serve as a solid baseline for future research. The code and data will be available at https://github.com/GangmingZhao/tb-attribute-weak-localization.
GREN: Graph-Regularized Embedding Network for Weakly-Supervised Disease Localization in X-ray images
Jul 14, 2021



Abstract:Locating diseases in chest X-ray images with few careful annotations saves large human effort. Recent works approached this task with innovative weakly-supervised algorithms such as multi-instance learning (MIL) and class activation maps (CAM), however, these methods often yield inaccurate or incomplete regions. One of the reasons is the neglection of the pathological implications hidden in the relationship across anatomical regions within each image and the relationship across images. In this paper, we argue that the cross-region and cross-image relationship, as contextual and compensating information, is vital to obtain more consistent and integral regions. To model the relationship, we propose the Graph Regularized Embedding Network (GREN), which leverages the intra-image and inter-image information to locate diseases on chest X-ray images. GREN uses a pre-trained U-Net to segment the lung lobes, and then models the intra-image relationship between the lung lobes using an intra-image graph to compare different regions. Meanwhile, the relationship between in-batch images is modeled by an inter-image graph to compare multiple images. This process mimics the training and decision-making process of a radiologist: comparing multiple regions and images for diagnosis. In order for the deep embedding layers of the neural network to retain structural information (important in the localization task), we use the Hash coding and Hamming distance to compute the graphs, which are used as regularizers to facilitate training. By means of this, our approach achieves the state-of-the-art result on NIH chest X-ray dataset for weakly-supervised disease localization. Our codes are accessible online.
Cross Chest Graph for Disease Diagnosis with Structural Relational Reasoning
Feb 01, 2021



Abstract:Locating lesions is important in the computer-aided diagnosis of X-ray images. However, box-level annotation is time-consuming and laborious. How to locate lesions accurately with few, or even without careful annotations is an urgent problem. Although several works have approached this problem with weakly-supervised methods, the performance needs to be improved. One obstacle is that general weakly-supervised methods have failed to consider the characteristics of X-ray images, such as the highly-structural attribute. We therefore propose the Cross-chest Graph (CCG), which improves the performance of automatic lesion detection by imitating doctor's training and decision-making process. CCG models the intra-image relationship between different anatomical areas by leveraging the structural information to simulate the doctor's habit of observing different areas. Meanwhile, the relationship between any pair of images is modeled by a knowledge-reasoning module to simulate the doctor's habit of comparing multiple images. We integrate intra-image and inter-image information into a unified end-to-end framework. Experimental results on the NIH Chest-14 database (112,120 frontal-view X-ray images with 14 diseases) demonstrate that the proposed method achieves state-of-the-art performance in weakly-supervised localization of lesions by absorbing professional knowledge in the medical field.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge