Tianyun Wang
Spatial Temporal Attention based Target Vehicle Trajectory Prediction for Internet of Vehicles
Jan 01, 2025



Abstract:Forecasting vehicle behavior within complex traffic environments is pivotal within Intelligent Transportation Systems (ITS). Though this technology plays a significant role in alleviating the prevalent operational difficulties in logistics and transportation systems, the precise prediction of vehicle trajectories still poses a substantial challenge. To address this, our study introduces the Spatio Temporal Attention-based methodology for Target Vehicle Trajectory Prediction (STATVTPred). This approach integrates Global Positioning System(GPS) localization technology to track target movement and dynamically predict the vehicle's future path using comprehensive spatio-temporal trajectory data. We map the vehicle trajectory onto a directed graph, after which spatial attributes are extracted via a Graph Attention Networks(GATs). The Transformer technology is employed to yield temporal features from the sequence. These elements are then amalgamated with local road network structure maps to filter and deliver a smooth trajectory sequence, resulting in precise vehicle trajectory prediction.This study validates our proposed STATVTPred method on T-Drive and Chengdu taxi-trajectory datasets. The experimental results demonstrate that STATVTPred achieves 6.38% and 10.55% higher Average Match Rate (AMR) than the Transformer model on the Beijing and Chengdu datasets, respectively. Compared to the LSTM Encoder-Decoder model, STATVTPred boosts AMR by 37.45% and 36.06% on the same datasets. This is expected to establish STATVTPred as a new approach for handling trajectory prediction of targets in logistics and transportation scenarios, thereby enhancing prediction accuracy.
A Survey for Large Language Models in Biomedicine
Aug 29, 2024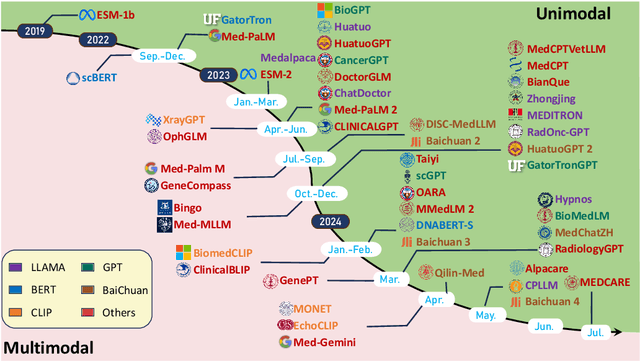
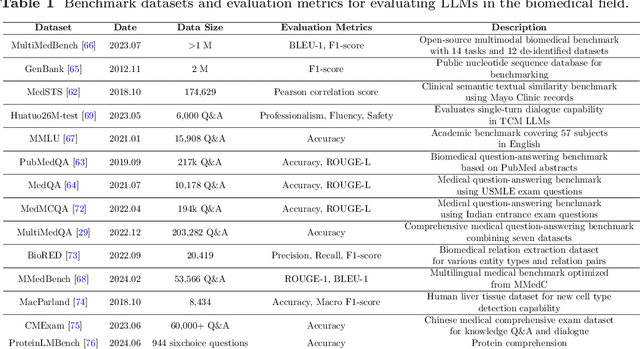
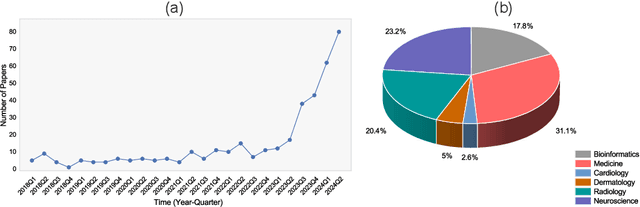
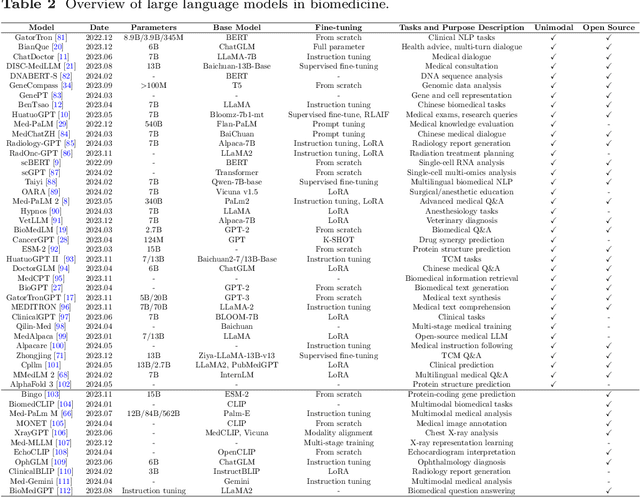
Abstract:Recent breakthroughs in large language models (LLMs) offer unprecedented natural language understanding and generation capabilities. However, existing surveys on LLMs in biomedicine often focus on specific applications or model architectures, lacking a comprehensive analysis that integrates the latest advancements across various biomedical domains. This review, based on an analysis of 484 publications sourced from databases including PubMed, Web of Science, and arXiv, provides an in-depth examination of the current landscape, applications, challenges, and prospects of LLMs in biomedicine, distinguishing itself by focusing on the practical implications of these models in real-world biomedical contexts. Firstly, we explore the capabilities of LLMs in zero-shot learning across a broad spectrum of biomedical tasks, including diagnostic assistance, drug discovery, and personalized medicine, among others, with insights drawn from 137 key studies. Then, we discuss adaptation strategies of LLMs, including fine-tuning methods for both uni-modal and multi-modal LLMs to enhance their performance in specialized biomedical contexts where zero-shot fails to achieve, such as medical question answering and efficient processing of biomedical literature. Finally, we discuss the challenges that LLMs face in the biomedicine domain including data privacy concerns, limited model interpretability, issues with dataset quality, and ethics due to the sensitive nature of biomedical data, the need for highly reliable model outputs, and the ethical implications of deploying AI in healthcare. To address these challenges, we also identify future research directions of LLM in biomedicine including federated learning methods to preserve data privacy and integrating explainable AI methodologies to enhance the transparency of LLMs.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge