Tianyi Qian
Venus: An Efficient Edge Memory-and-Retrieval System for VLM-based Online Video Understanding
Dec 08, 2025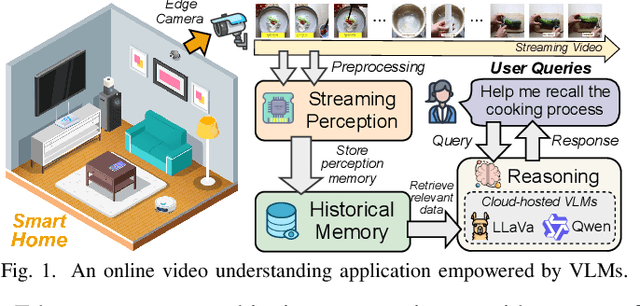
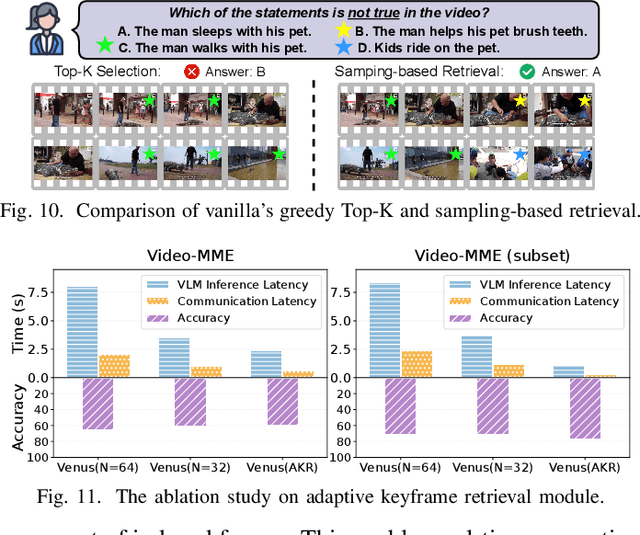
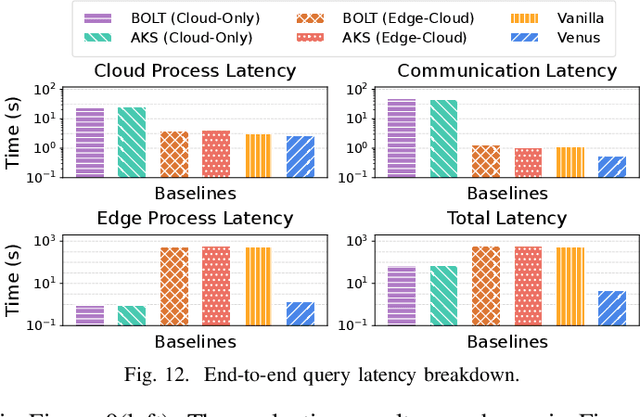
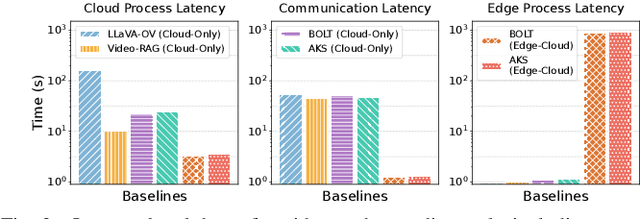
Abstract:Vision-language models (VLMs) have demonstrated impressive multimodal comprehension capabilities and are being deployed in an increasing number of online video understanding applications. While recent efforts extensively explore advancing VLMs' reasoning power in these cases, deployment constraints are overlooked, leading to overwhelming system overhead in real-world deployments. To address that, we propose Venus, an on-device memory-and-retrieval system for efficient online video understanding. Venus proposes an edge-cloud disaggregated architecture that sinks memory construction and keyframe retrieval from cloud to edge, operating in two stages. In the ingestion stage, Venus continuously processes streaming edge videos via scene segmentation and clustering, where the selected keyframes are embedded with a multimodal embedding model to build a hierarchical memory for efficient storage and retrieval. In the querying stage, Venus indexes incoming queries from memory, and employs a threshold-based progressive sampling algorithm for keyframe selection that enhances diversity and adaptively balances system cost and reasoning accuracy. Our extensive evaluation shows that Venus achieves a 15x-131x speedup in total response latency compared to state-of-the-art methods, enabling real-time responses within seconds while maintaining comparable or even superior reasoning accuracy.
Jupiter: Fast and Resource-Efficient Collaborative Inference of Generative LLMs on Edge Devices
Apr 11, 2025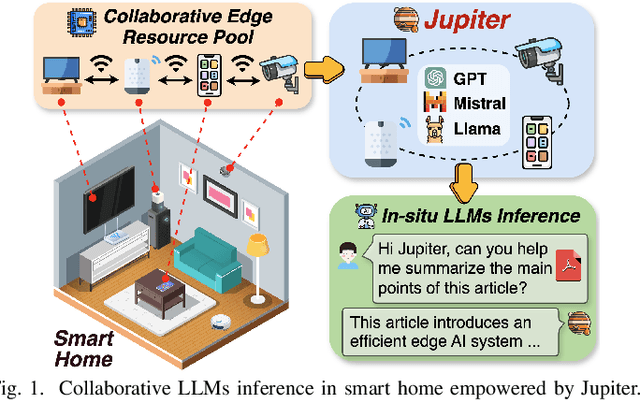
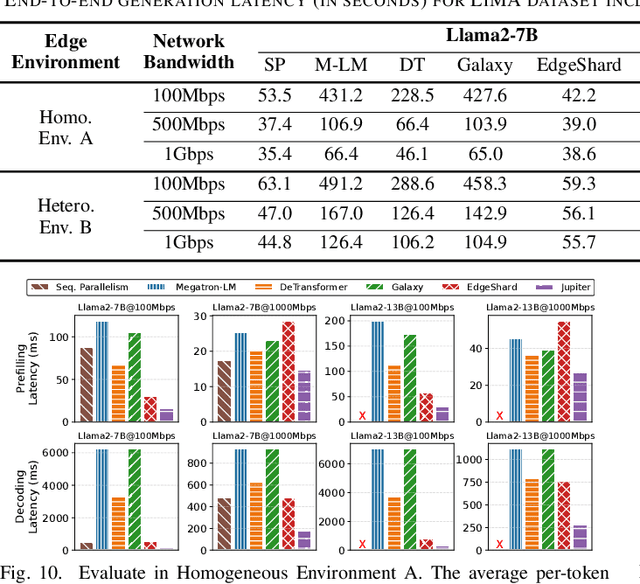
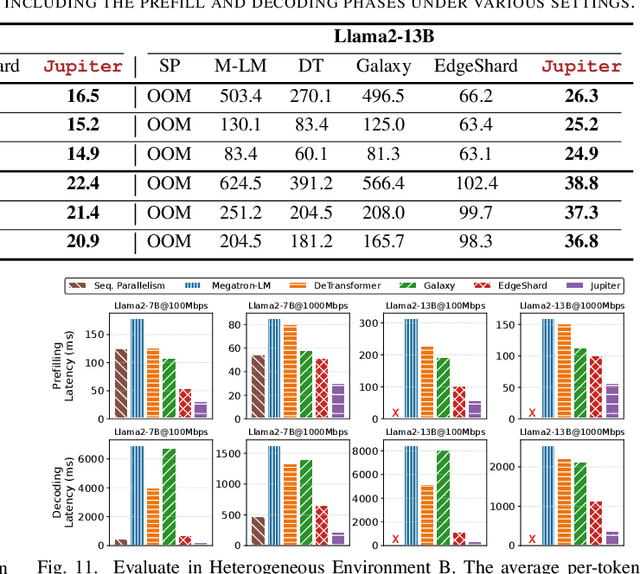
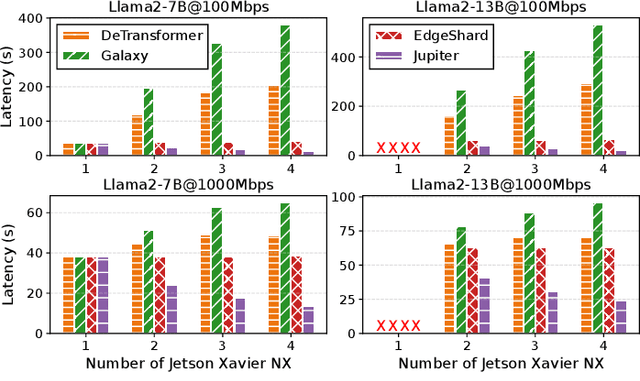
Abstract:Generative large language models (LLMs) have garnered significant attention due to their exceptional capabilities in various AI tasks. Traditionally deployed in cloud datacenters, LLMs are now increasingly moving towards more accessible edge platforms to protect sensitive user data and ensure privacy preservation. The limited computational resources of individual edge devices, however, can result in excessively prolonged inference latency and overwhelmed memory usage. While existing research has explored collaborative edge computing to break the resource wall of individual devices, these solutions yet suffer from massive communication overhead and under-utilization of edge resources. Furthermore, they focus exclusively on optimizing the prefill phase, neglecting the crucial autoregressive decoding phase for generative LLMs. To address that, we propose Jupiter, a fast, scalable, and resource-efficient collaborative edge AI system for generative LLM inference. Jupiter introduces a flexible pipelined architecture as a principle and differentiates its system design according to the differentiated characteristics of the prefill and decoding phases. For prefill phase, Jupiter submits a novel intra-sequence pipeline parallelism and develops a meticulous parallelism planning strategy to maximize resource efficiency; For decoding, Jupiter devises an effective outline-based pipeline parallel decoding mechanism combined with speculative decoding, which further magnifies inference acceleration. Extensive evaluation based on realistic implementation demonstrates that Jupiter remarkably outperforms state-of-the-art approaches under various edge environment setups, achieving up to 26.1x end-to-end latency reduction while rendering on-par generation quality.
Prompt-Guided Dual-Path UNet with Mamba for Medical Image Segmentation
Mar 25, 2025Abstract:Convolutional neural networks (CNNs) and transformers are widely employed in constructing UNet architectures for medical image segmentation tasks. However, CNNs struggle to model long-range dependencies, while transformers suffer from quadratic computational complexity. Recently, Mamba, a type of State Space Models, has gained attention for its exceptional ability to model long-range interactions while maintaining linear computational complexity. Despite the emergence of several Mamba-based methods, they still present the following limitations: first, their network designs generally lack perceptual capabilities for the original input data; second, they primarily focus on capturing global information, while often neglecting local details. To address these challenges, we propose a prompt-guided CNN-Mamba dual-path UNet, termed PGM-UNet, for medical image segmentation. Specifically, we introduce a prompt-guided residual Mamba module that adaptively extracts dynamic visual prompts from the original input data, effectively guiding Mamba in capturing global information. Additionally, we design a local-global information fusion network, comprising a local information extraction module, a prompt-guided residual Mamba module, and a multi-focus attention fusion module, which effectively integrates local and global information. Furthermore, inspired by Kolmogorov-Arnold Networks (KANs), we develop a multi-scale information extraction module to capture richer contextual information without altering the resolution. We conduct extensive experiments on the ISIC-2017, ISIC-2018, DIAS, and DRIVE. The results demonstrate that the proposed method significantly outperforms state-of-the-art approaches in multiple medical image segmentation tasks.
Pluto and Charon: A Time and Memory Efficient Collaborative Edge AI Framework for Personal LLMs Fine-Tuning
Aug 20, 2024Abstract:Large language models (LLMs) have unlocked a plethora of powerful applications at the network edge, such as intelligent personal assistants. Data privacy and security concerns have prompted a shift towards edge-based fine-tuning of personal LLMs, away from cloud reliance. However, this raises issues of computational intensity and resource scarcity, hindering training efficiency and feasibility. While current studies investigate parameter-efficient fine-tuning (PEFT) techniques to mitigate resource constraints, our analysis indicates that these techniques are not sufficiently resource-efficient for edge devices. To tackle these challenges, we propose Pluto and Charon (PAC), a time and memory efficient collaborative edge AI framework for personal LLMs fine-tuning. PAC breaks the resource wall of personal LLMs fine-tuning with a sophisticated algorithm-system co-design. (1) Algorithmically, PAC implements a personal LLMs fine-tuning technique that is efficient in terms of parameters, time, and memory. It utilizes Parallel Adapters to circumvent the need for a full backward pass through the LLM backbone. Additionally, an activation cache mechanism further streamlining the process by negating the necessity for repeated forward passes across multiple epochs. (2) Systematically, PAC leverages edge devices in close proximity, pooling them as a collective resource for in-situ personal LLMs fine-tuning, utilizing a hybrid data and pipeline parallelism to orchestrate distributed training. The use of the activation cache eliminates the need for forward pass through the LLM backbone,enabling exclusive fine-tuning of the Parallel Adapters using data parallelism. Extensive evaluation based on prototype implementation demonstrates that PAC remarkably outperforms state-of-the-art approaches, achieving up to 8.64x end-to-end speedup and up to 88.16% reduction in memory footprint.
Deep Convolutional Neural Networks for Molecular Subtyping of Gliomas Using Magnetic Resonance Imaging
Mar 10, 2022Abstract:Knowledge of molecular subtypes of gliomas can provide valuable information for tailored therapies. This study aimed to investigate the use of deep convolutional neural networks (DCNNs) for noninvasive glioma subtyping with radiological imaging data according to the new taxonomy announced by the World Health Organization in 2016. Methods: A DCNN model was developed for the prediction of the five glioma subtypes based on a hierarchical classification paradigm. This model used three parallel, weight-sharing, deep residual learning networks to process 2.5-dimensional input of trimodal MRI data, including T1-weighted, T1-weighted with contrast enhancement, and T2-weighted images. A data set comprising 1,016 real patients was collected for evaluation of the developed DCNN model. The predictive performance was evaluated via the area under the curve (AUC) from the receiver operating characteristic analysis. For comparison, the performance of a radiomics-based approach was also evaluated. Results: The AUCs of the DCNN model for the four classification tasks in the hierarchical classification paradigm were 0.89, 0.89, 0.85, and 0.66, respectively, as compared to 0.85, 0.75, 0.67, and 0.59 of the radiomics approach. Conclusion: The results showed that the developed DCNN model can predict glioma subtypes with promising performance, given sufficient, non-ill-balanced training data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge