Yinyan Wang
Translation, Scale and Rotation: Cross-Modal Alignment Meets RGB-Infrared Vehicle Detection
Sep 28, 2022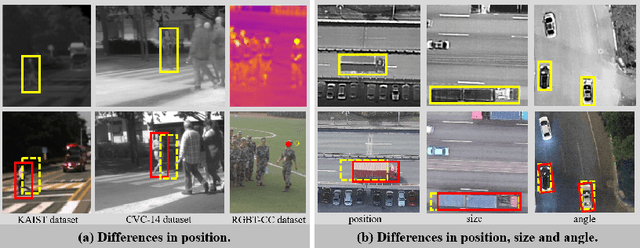


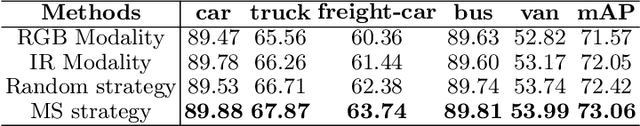
Abstract:Integrating multispectral data in object detection, especially visible and infrared images, has received great attention in recent years. Since visible (RGB) and infrared (IR) images can provide complementary information to handle light variations, the paired images are used in many fields, such as multispectral pedestrian detection, RGB-IR crowd counting and RGB-IR salient object detection. Compared with natural RGB-IR images, we find detection in aerial RGB-IR images suffers from cross-modal weakly misalignment problems, which are manifested in the position, size and angle deviations of the same object. In this paper, we mainly address the challenge of cross-modal weakly misalignment in aerial RGB-IR images. Specifically, we firstly explain and analyze the cause of the weakly misalignment problem. Then, we propose a Translation-Scale-Rotation Alignment (TSRA) module to address the problem by calibrating the feature maps from these two modalities. The module predicts the deviation between two modality objects through an alignment process and utilizes Modality-Selection (MS) strategy to improve the performance of alignment. Finally, a two-stream feature alignment detector (TSFADet) based on the TSRA module is constructed for RGB-IR object detection in aerial images. With comprehensive experiments on the public DroneVehicle datasets, we verify that our method reduces the effect of the cross-modal misalignment and achieve robust detection results.
Deep Convolutional Neural Networks for Molecular Subtyping of Gliomas Using Magnetic Resonance Imaging
Mar 10, 2022Abstract:Knowledge of molecular subtypes of gliomas can provide valuable information for tailored therapies. This study aimed to investigate the use of deep convolutional neural networks (DCNNs) for noninvasive glioma subtyping with radiological imaging data according to the new taxonomy announced by the World Health Organization in 2016. Methods: A DCNN model was developed for the prediction of the five glioma subtypes based on a hierarchical classification paradigm. This model used three parallel, weight-sharing, deep residual learning networks to process 2.5-dimensional input of trimodal MRI data, including T1-weighted, T1-weighted with contrast enhancement, and T2-weighted images. A data set comprising 1,016 real patients was collected for evaluation of the developed DCNN model. The predictive performance was evaluated via the area under the curve (AUC) from the receiver operating characteristic analysis. For comparison, the performance of a radiomics-based approach was also evaluated. Results: The AUCs of the DCNN model for the four classification tasks in the hierarchical classification paradigm were 0.89, 0.89, 0.85, and 0.66, respectively, as compared to 0.85, 0.75, 0.67, and 0.59 of the radiomics approach. Conclusion: The results showed that the developed DCNN model can predict glioma subtypes with promising performance, given sufficient, non-ill-balanced training data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge