Sylwia Majchrowska
Unlocking the Heart Using Adaptive Locked Agnostic Networks
Sep 21, 2023



Abstract:Supervised training of deep learning models for medical imaging applications requires a significant amount of labeled data. This is posing a challenge as the images are required to be annotated by medical professionals. To address this limitation, we introduce the Adaptive Locked Agnostic Network (ALAN), a concept involving self-supervised visual feature extraction using a large backbone model to produce anatomically robust semantic self-segmentation. In the ALAN methodology, this self-supervised training occurs only once on a large and diverse dataset. Due to the intuitive interpretability of the segmentation, downstream models tailored for specific tasks can be easily designed using white-box models with few parameters. This, in turn, opens up the possibility of communicating the inner workings of a model with domain experts and introducing prior knowledge into it. It also means that the downstream models become less data-hungry compared to fully supervised approaches. These characteristics make ALAN particularly well-suited for resource-scarce scenarios, such as costly clinical trials and rare diseases. In this paper, we apply the ALAN approach to three publicly available echocardiography datasets: EchoNet-Dynamic, CAMUS, and TMED-2. Our findings demonstrate that the self-supervised backbone model robustly identifies anatomical subregions of the heart in an apical four-chamber view. Building upon this, we design two downstream models, one for segmenting a target anatomical region, and a second for echocardiogram view classification.
Towards Explainable Motion Prediction using Heterogeneous Graph Representations
Dec 07, 2022Abstract:Motion prediction systems aim to capture the future behavior of traffic scenarios enabling autonomous vehicles to perform safe and efficient planning. The evolution of these scenarios is highly uncertain and depends on the interactions of agents with static and dynamic objects in the scene. GNN-based approaches have recently gained attention as they are well suited to naturally model these interactions. However, one of the main challenges that remains unexplored is how to address the complexity and opacity of these models in order to deal with the transparency requirements for autonomous driving systems, which includes aspects such as interpretability and explainability. In this work, we aim to improve the explainability of motion prediction systems by using different approaches. First, we propose a new Explainable Heterogeneous Graph-based Policy (XHGP) model based on an heterograph representation of the traffic scene and lane-graph traversals, which learns interaction behaviors using object-level and type-level attention. This learned attention provides information about the most important agents and interactions in the scene. Second, we explore this same idea with the explanations provided by GNNExplainer. Third, we apply counterfactual reasoning to provide explanations of selected individual scenarios by exploring the sensitivity of the trained model to changes made to the input data, i.e., masking some elements of the scene, modifying trajectories, and adding or removing dynamic agents. The explainability analysis provided in this paper is a first step towards more transparent and reliable motion prediction systems, important from the perspective of the user, developers and regulatory agencies. The code to reproduce this work is publicly available at https://github.com/sancarlim/Explainable-MP/tree/v1.1.
Towards Trustworthy Multi-Modal Motion Prediction: Evaluation and Interpretability
Oct 28, 2022



Abstract:Predicting the motion of other road agents enables autonomous vehicles to perform safe and efficient path planning. This task is very complex, as the behaviour of road agents depends on many factors and the number of possible future trajectories can be considerable (multi-modal). Most approaches proposed to address multi-modal motion prediction are based on complex machine learning systems that have limited interpretability. Moreover, the metrics used in current benchmarks do not evaluate all aspects of the problem, such as the diversity and admissibility of the output. In this work, we aim to advance towards the design of trustworthy motion prediction systems, based on some of the requirements for the design of Trustworthy Artificial Intelligence. We focus on evaluation criteria, robustness, and interpretability of outputs. First, we comprehensively analyse the evaluation metrics, identify the main gaps of current benchmarks, and propose a new holistic evaluation framework. In addition, we formulate a method for the assessment of spatial and temporal robustness by simulating noise in the perception system. We propose an intent prediction layer that can be attached to multi-modal motion prediction models to enhance the interpretability of the outputs and generate more balanced results in the proposed evaluation framework. Finally, the interpretability of the outputs is assessed by means of a survey that explores different elements in the visualization of the multi-modal trajectories and intentions.
GAN-based generative modelling for dermatological applications -- comparative study
Aug 24, 2022



Abstract:The lack of sufficiently large open medical databases is one of the biggest challenges in AI-powered healthcare. Synthetic data created using Generative Adversarial Networks (GANs) appears to be a good solution to mitigate the issues with privacy policies. The other type of cure is decentralized protocol across multiple medical institutions without exchanging local data samples. In this paper, we explored unconditional and conditional GANs in centralized and decentralized settings. The centralized setting imitates studies on large but highly unbalanced skin lesion dataset, while the decentralized one simulates a more realistic hospital scenario with three institutions. We evaluated models' performance in terms of fidelity, diversity, speed of training, and predictive ability of classifiers trained on the generated synthetic data. In addition we provided explainability through exploration of latent space and embeddings projection focused both on global and local explanations. Calculated distance between real images and their projections in the latent space proved the authenticity and generalization of trained GANs, which is one of the main concerns in this type of applications. The open source code for conducted studies is publicly available at \url{https://github.com/aidotse/stylegan2-ada-pytorch}.
The biasing effect of GAN-based augmentation methods on skin lesion images
Jun 30, 2022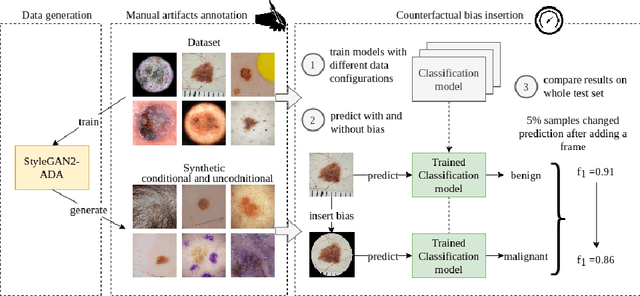
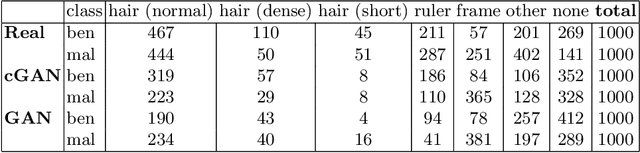
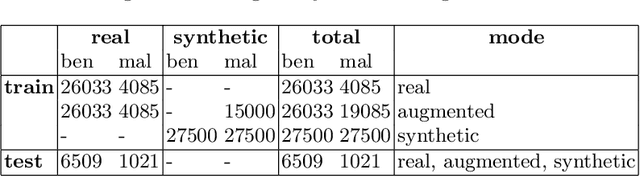

Abstract:New medical datasets are now more open to the public, allowing for better and more extensive research. Although prepared with the utmost care, new datasets might still be a source of spurious correlations that affect the learning process. Moreover, data collections are usually not large enough and are often unbalanced. One approach to alleviate the data imbalance is using data augmentation with Generative Adversarial Networks (GANs) to extend the dataset with high-quality images. GANs are usually trained on the same biased datasets as the target data, resulting in more biased instances. This work explored unconditional and conditional GANs to compare their bias inheritance and how the synthetic data influenced the models. We provided extensive manual data annotation of possibly biasing artifacts on the well-known ISIC dataset with skin lesions. In addition, we examined classification models trained on both real and synthetic data with counterfactual bias explanations. Our experiments showed that GANs inherited biases and sometimes even amplified them, leading to even stronger spurious correlations. Manual data annotation and synthetic images are publicly available for reproducible scientific research.
Open Source HamNoSys Parser for Multilingual Sign Language Encoding
Apr 14, 2022



Abstract:This paper presents our recent developments in the field of automatic processing of sign language corpora using the Hamburg Sign Language Annotation System (HamNoSys). We designed an automated tool to convert HamNoSys annotations into numerical labels for defined initial features of body and hand positions. Our proposed numerical multilabels greatly simplify the structure of HamNoSys annotation without significant loss of gloss meaning. These numerical multilabels can potentially be used to feed the machine learning models, which would accelerate the development of vision-based sign language recognition. In addition, this tool can assist experts in the annotation process to help identify semantic errors. The code and sample annotations are publicly available at https://github.com/hearai/parse-hamnosys.
Self-Normalized Density Map (SNDM) for Counting Microbiological Objects
Mar 15, 2022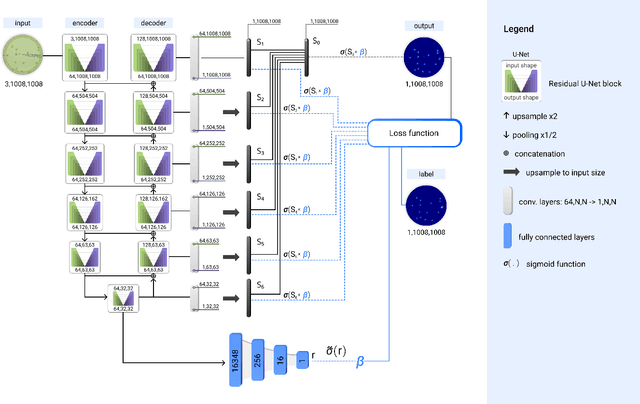
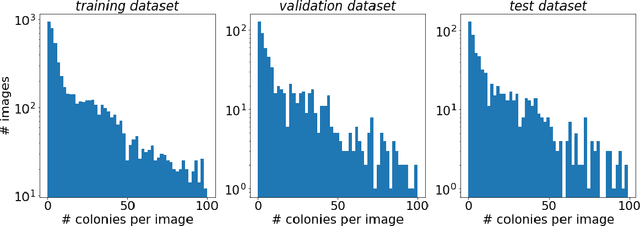
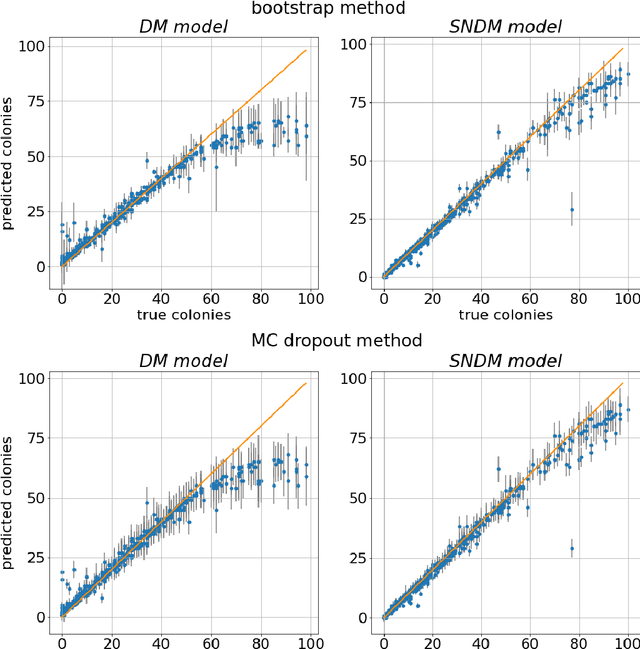
Abstract:The statistical properties of the density map (DM) approach to counting microbiological objects on images are studied in detail. The DM is given by U$^2$-Net. Two statistical methods for deep neural networks are utilized: the bootstrap and the Monte Carlo (MC) dropout. The detailed analysis of the uncertainties for the DM predictions leads to a deeper understanding of the DM model's deficiencies. Based on our investigation, we propose a self-normalization module in the network. The improved network model, called Self-Normalized Density Map (SNDM), can correct its output density map by itself to accurately predict the total number of objects in the image. The SNDM architecture outperforms the original model. Moreover, both statistical frameworks -- bootstrap and MC dropout -- have consistent statistical results for SNDM, which were not observed in the original model.
Generation of microbial colonies dataset with deep learning style transfer
Nov 06, 2021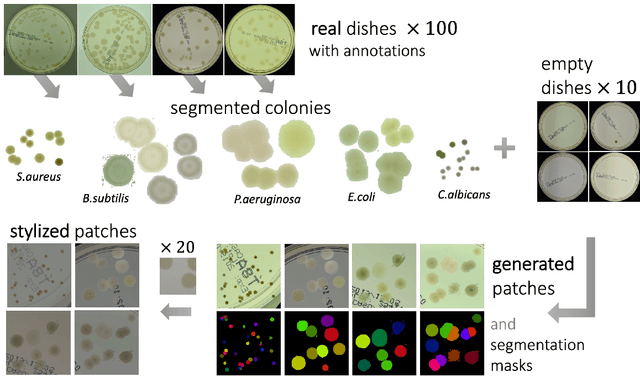
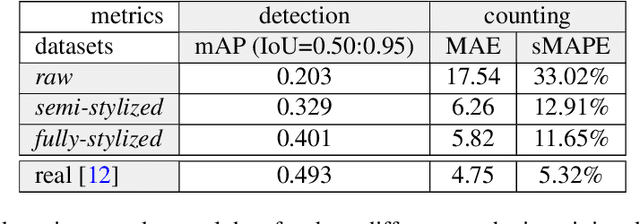
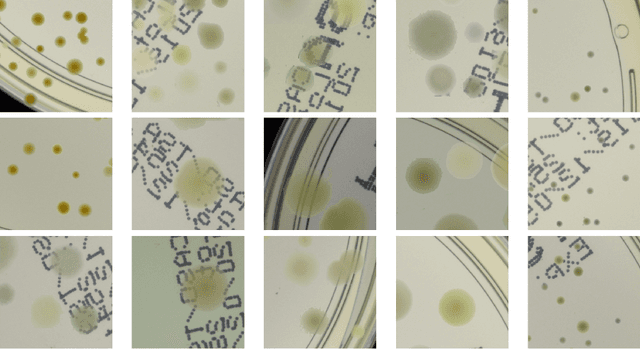
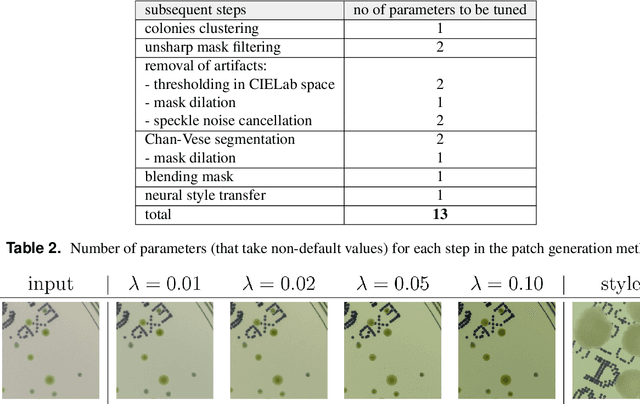
Abstract:We introduce an effective strategy to generate a synthetic dataset of microbiological images of Petri dishes that can be used to train deep learning models. The developed generator employs traditional computer vision algorithms together with a neural style transfer method for data augmentation. We show that the method is able to synthesize a dataset of realistic looking images that can be used to train a neural network model capable of localising, segmenting, and classifying five different microbial species. Our method requires significantly fewer resources to obtain a useful dataset than collecting and labeling a whole large set of real images with annotations. We show that starting with only 100 real images, we can generate data to train a detector that achieves comparable results to the same detector but trained on a real, several dozen times bigger dataset. We prove the usefulness of the method in microbe detection and segmentation, but we expect that it is general and flexible and can also be applicable in other domains of science and industry to detect various objects.
Deep neural networks approach to microbial colony detection -- a comparative analysis
Aug 24, 2021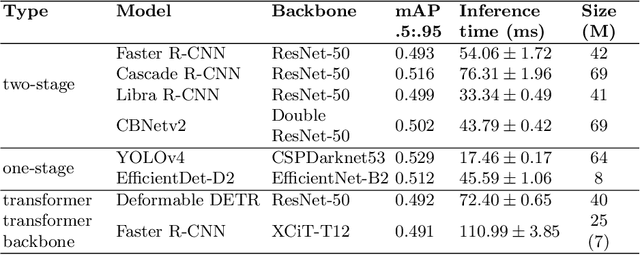
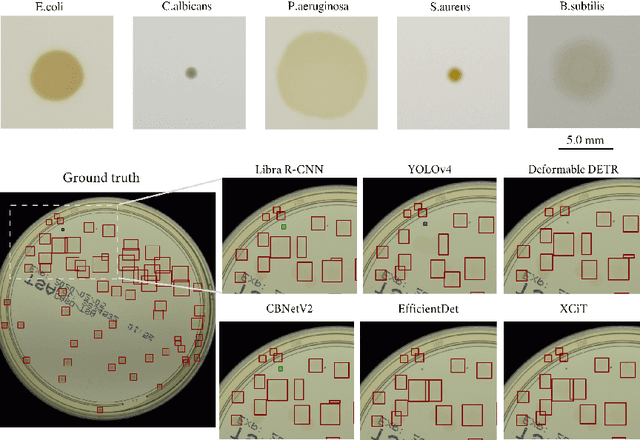
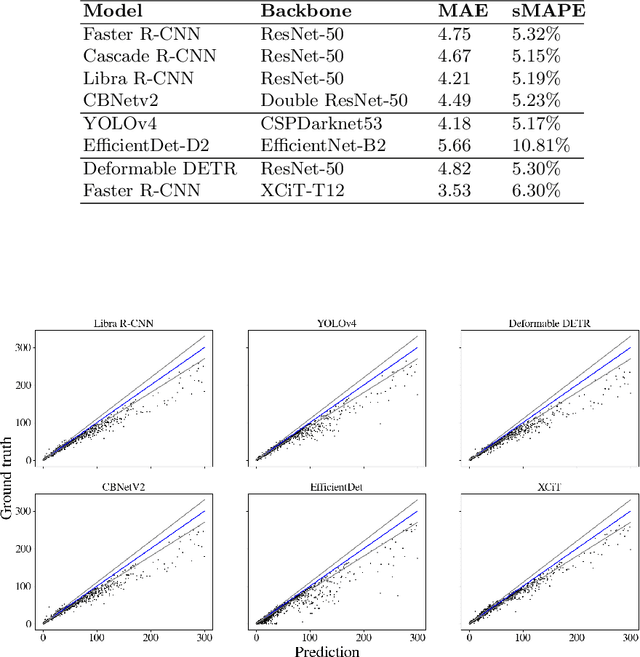
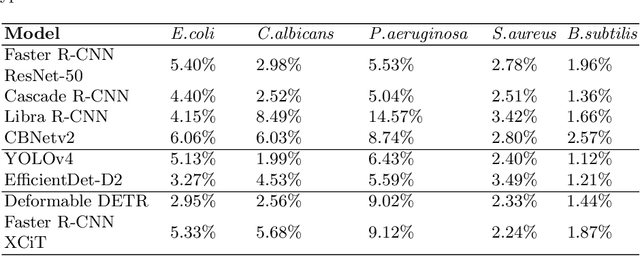
Abstract:Counting microbial colonies is a fundamental task in microbiology and has many applications in numerous industry branches. Despite this, current studies towards automatic microbial counting using artificial intelligence are hardly comparable due to the lack of unified methodology and the availability of large datasets. The recently introduced AGAR dataset is the answer to the second need, but the research carried out is still not exhaustive. To tackle this problem, we compared the performance of three well-known deep learning approaches for object detection on the AGAR dataset, namely two-stage, one-stage and transformer based neural networks. The achieved results may serve as a benchmark for future experiments.
AGAR a microbial colony dataset for deep learning detection
Aug 03, 2021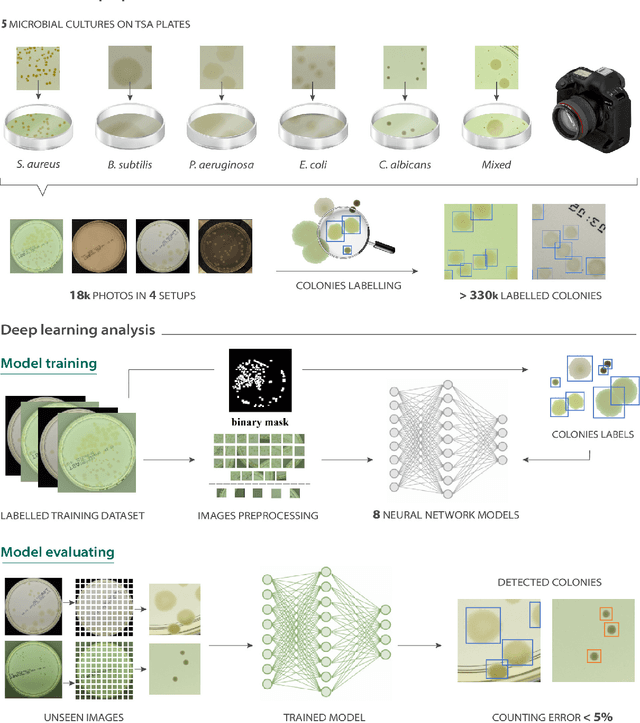
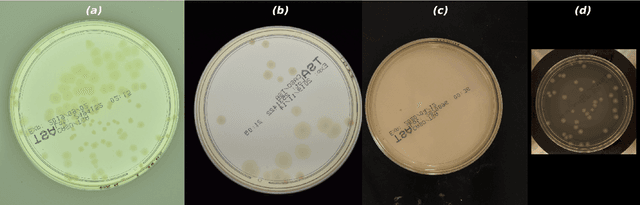
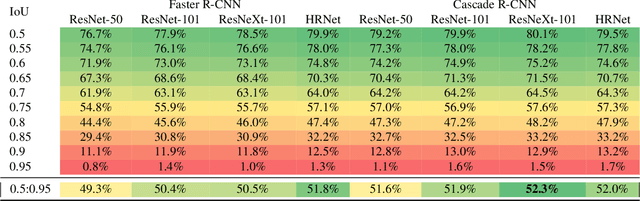
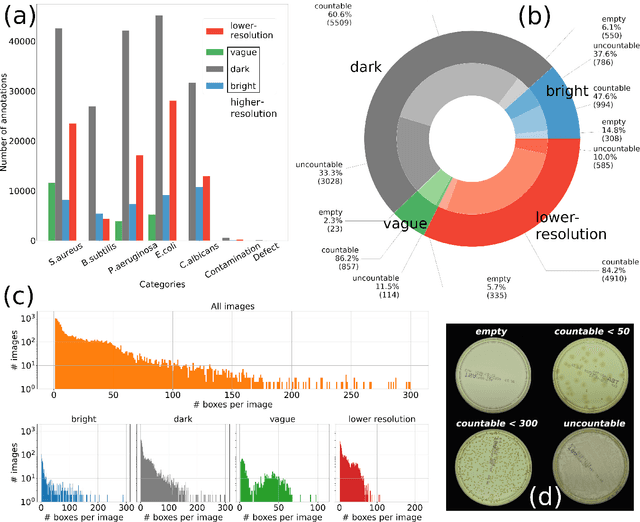
Abstract:The Annotated Germs for Automated Recognition (AGAR) dataset is an image database of microbial colonies cultured on agar plates. It contains 18000 photos of five different microorganisms as single or mixed cultures, taken under diverse lighting conditions with two different cameras. All the images are classified into "countable", "uncountable", and "empty", with the "countable" class labeled by microbiologists with colony location and species identification (336442 colonies in total). This study describes the dataset itself and the process of its development. In the second part, the performance of selected deep neural network architectures for object detection, namely Faster R-CNN and Cascade R-CNN, was evaluated on the AGAR dataset. The results confirmed the great potential of deep learning methods to automate the process of microbe localization and classification based on Petri dish photos. Moreover, AGAR is the first publicly available dataset of this kind and size and will facilitate the future development of machine learning models. The data used in these studies can be found at https://agar.neurosys.com/.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge