Anders Hildeman
Unlocking the Heart Using Adaptive Locked Agnostic Networks
Sep 21, 2023



Abstract:Supervised training of deep learning models for medical imaging applications requires a significant amount of labeled data. This is posing a challenge as the images are required to be annotated by medical professionals. To address this limitation, we introduce the Adaptive Locked Agnostic Network (ALAN), a concept involving self-supervised visual feature extraction using a large backbone model to produce anatomically robust semantic self-segmentation. In the ALAN methodology, this self-supervised training occurs only once on a large and diverse dataset. Due to the intuitive interpretability of the segmentation, downstream models tailored for specific tasks can be easily designed using white-box models with few parameters. This, in turn, opens up the possibility of communicating the inner workings of a model with domain experts and introducing prior knowledge into it. It also means that the downstream models become less data-hungry compared to fully supervised approaches. These characteristics make ALAN particularly well-suited for resource-scarce scenarios, such as costly clinical trials and rare diseases. In this paper, we apply the ALAN approach to three publicly available echocardiography datasets: EchoNet-Dynamic, CAMUS, and TMED-2. Our findings demonstrate that the self-supervised backbone model robustly identifies anatomical subregions of the heart in an apical four-chamber view. Building upon this, we design two downstream models, one for segmenting a target anatomical region, and a second for echocardiogram view classification.
Whole-brain substitute CT generation using Markov random field mixture models
Sep 28, 2016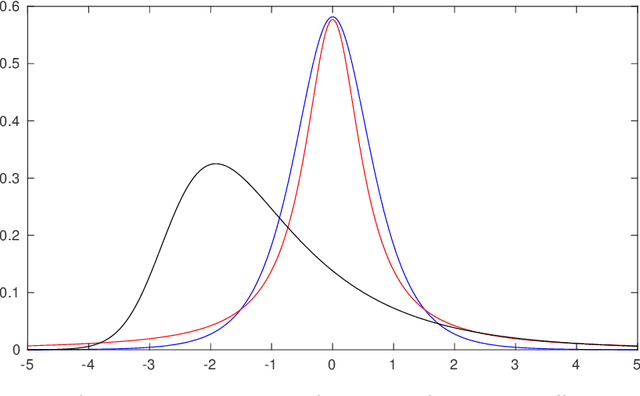
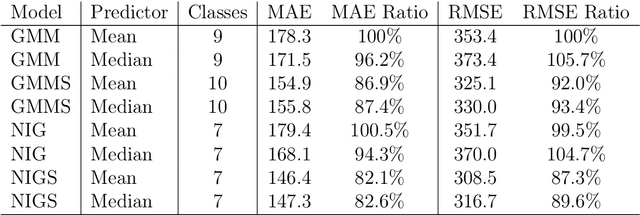
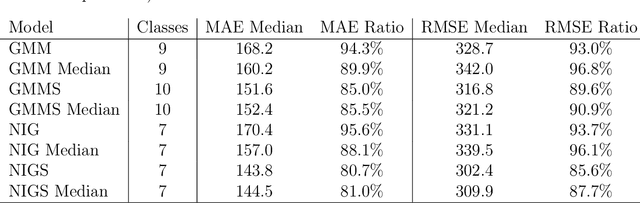
Abstract:Computed tomography (CT) equivalent information is needed for attenuation correction in PET imaging and for dose planning in radiotherapy. Prior work has shown that Gaussian mixture models can be used to generate a substitute CT (s-CT) image from a specific set of MRI modalities. This work introduces a more flexible class of mixture models for s-CT generation, that incorporates spatial dependency in the data through a Markov random field prior on the latent field of class memberships associated with a mixture model. Furthermore, the mixture distributions are extended from Gaussian to normal inverse Gaussian (NIG), allowing heavier tails and skewness. The amount of data needed to train a model for s-CT generation is of the order of 100 million voxels. The computational efficiency of the parameter estimation and prediction methods are hence paramount, especially when spatial dependency is included in the models. A stochastic Expectation Maximization (EM) gradient algorithm is proposed in order to tackle this challenge. The advantages of the spatial model and NIG distributions are evaluated with a cross-validation study based on data from 14 patients. The study show that the proposed model enhances the predictive quality of the s-CT images by reducing the mean absolute error with 17.9%. Also, the distribution of CT values conditioned on the MR images are better explained by the proposed model as evaluated using continuous ranked probability scores.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge