Tomasz Golan
Self-Normalized Density Map (SNDM) for Counting Microbiological Objects
Mar 15, 2022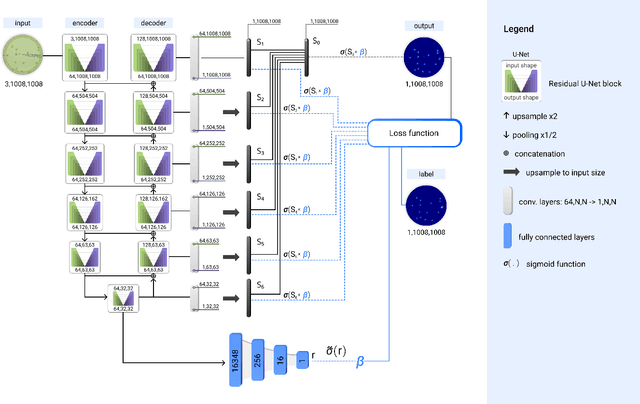
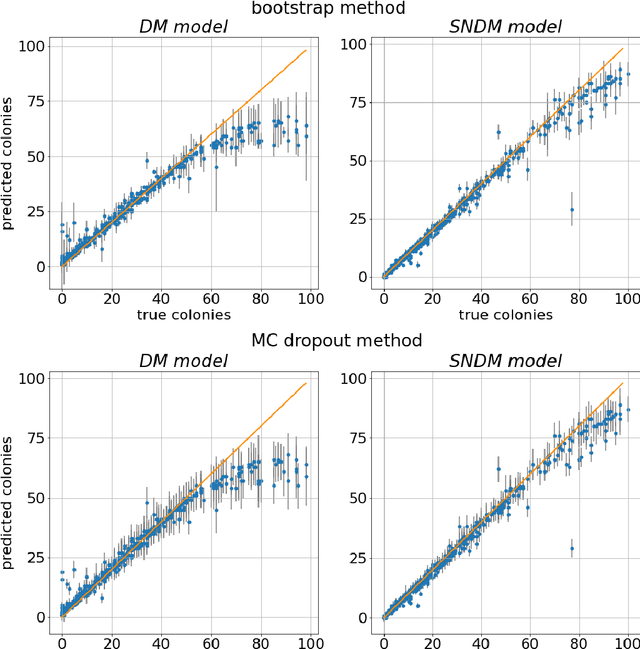
Abstract:The statistical properties of the density map (DM) approach to counting microbiological objects on images are studied in detail. The DM is given by U$^2$-Net. Two statistical methods for deep neural networks are utilized: the bootstrap and the Monte Carlo (MC) dropout. The detailed analysis of the uncertainties for the DM predictions leads to a deeper understanding of the DM model's deficiencies. Based on our investigation, we propose a self-normalization module in the network. The improved network model, called Self-Normalized Density Map (SNDM), can correct its output density map by itself to accurately predict the total number of objects in the image. The SNDM architecture outperforms the original model. Moreover, both statistical frameworks -- bootstrap and MC dropout -- have consistent statistical results for SNDM, which were not observed in the original model.
Generation of microbial colonies dataset with deep learning style transfer
Nov 06, 2021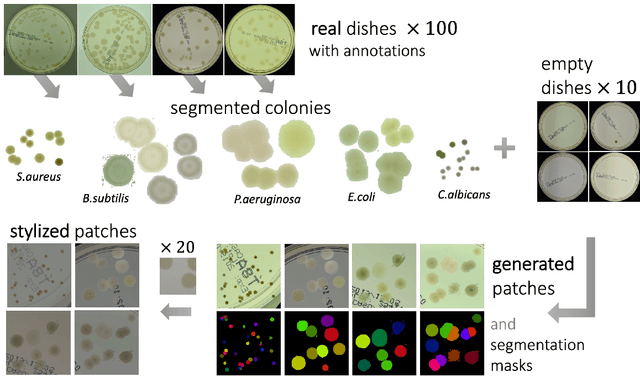
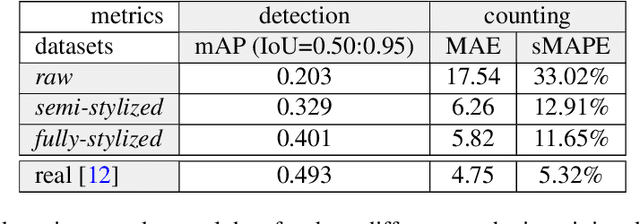
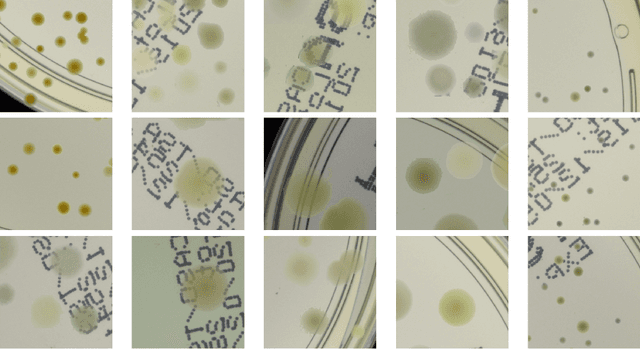
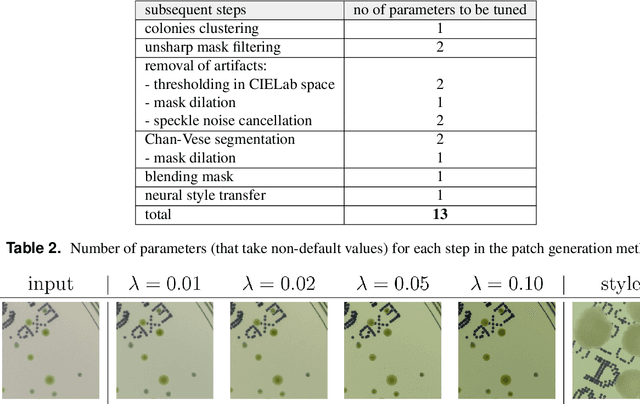
Abstract:We introduce an effective strategy to generate a synthetic dataset of microbiological images of Petri dishes that can be used to train deep learning models. The developed generator employs traditional computer vision algorithms together with a neural style transfer method for data augmentation. We show that the method is able to synthesize a dataset of realistic looking images that can be used to train a neural network model capable of localising, segmenting, and classifying five different microbial species. Our method requires significantly fewer resources to obtain a useful dataset than collecting and labeling a whole large set of real images with annotations. We show that starting with only 100 real images, we can generate data to train a detector that achieves comparable results to the same detector but trained on a real, several dozen times bigger dataset. We prove the usefulness of the method in microbe detection and segmentation, but we expect that it is general and flexible and can also be applicable in other domains of science and industry to detect various objects.
Deep neural networks approach to microbial colony detection -- a comparative analysis
Aug 24, 2021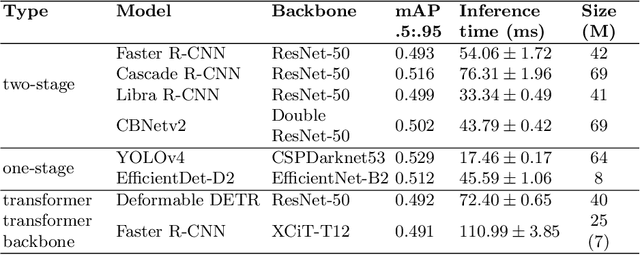
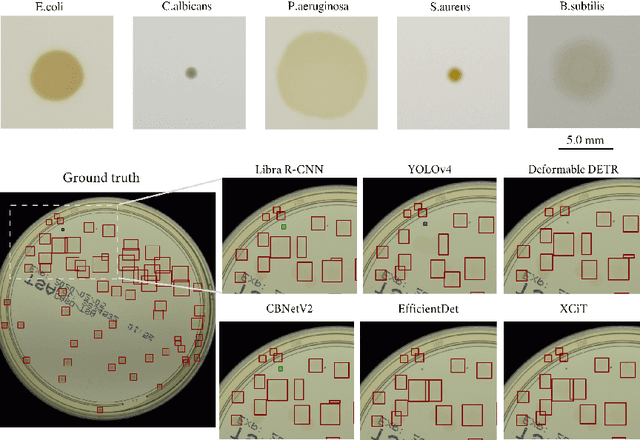
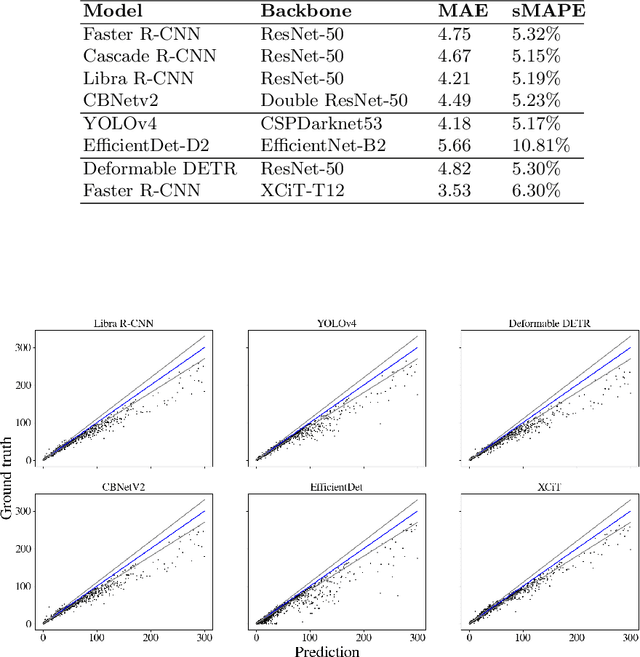
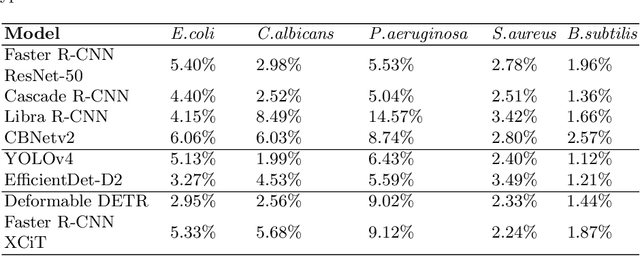
Abstract:Counting microbial colonies is a fundamental task in microbiology and has many applications in numerous industry branches. Despite this, current studies towards automatic microbial counting using artificial intelligence are hardly comparable due to the lack of unified methodology and the availability of large datasets. The recently introduced AGAR dataset is the answer to the second need, but the research carried out is still not exhaustive. To tackle this problem, we compared the performance of three well-known deep learning approaches for object detection on the AGAR dataset, namely two-stage, one-stage and transformer based neural networks. The achieved results may serve as a benchmark for future experiments.
AGAR a microbial colony dataset for deep learning detection
Aug 03, 2021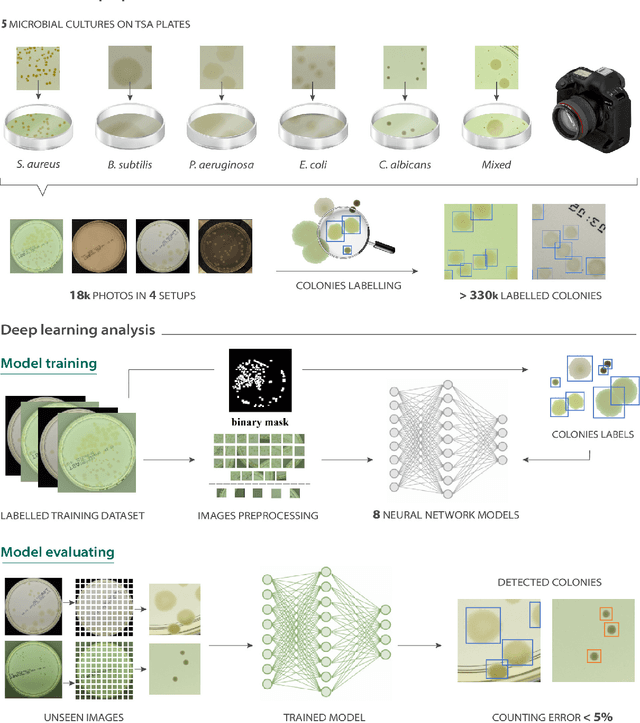
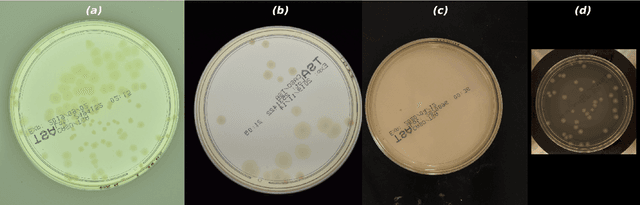
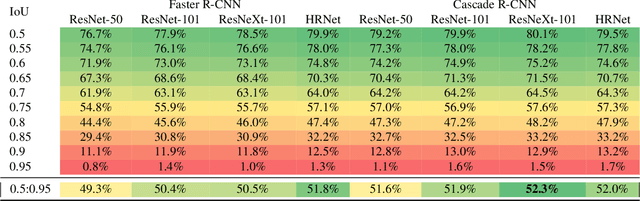
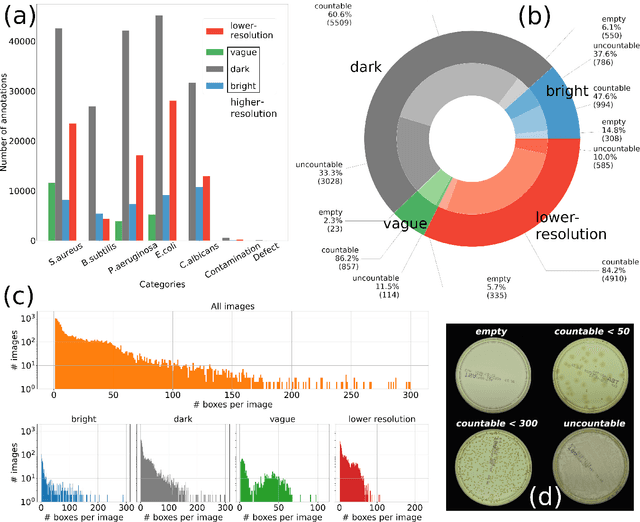
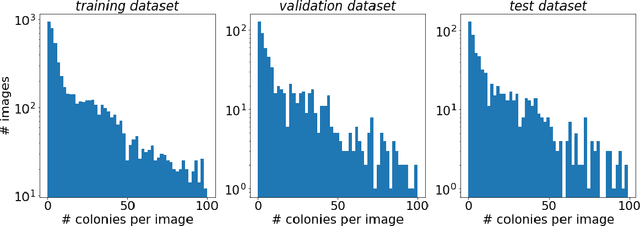
Abstract:The Annotated Germs for Automated Recognition (AGAR) dataset is an image database of microbial colonies cultured on agar plates. It contains 18000 photos of five different microorganisms as single or mixed cultures, taken under diverse lighting conditions with two different cameras. All the images are classified into "countable", "uncountable", and "empty", with the "countable" class labeled by microbiologists with colony location and species identification (336442 colonies in total). This study describes the dataset itself and the process of its development. In the second part, the performance of selected deep neural network architectures for object detection, namely Faster R-CNN and Cascade R-CNN, was evaluated on the AGAR dataset. The results confirmed the great potential of deep learning methods to automate the process of microbe localization and classification based on Petri dish photos. Moreover, AGAR is the first publicly available dataset of this kind and size and will facilitate the future development of machine learning models. The data used in these studies can be found at https://agar.neurosys.com/.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge