Jarosław Pawłowski
AI-enhanced tuning of quantum dot Hamiltonians toward Majorana modes
Jan 05, 2026Abstract:We propose a neural network-based model capable of learning the broad landscape of working regimes in quantum dot simulators, and using this knowledge to autotune these devices - based on transport measurements - toward obtaining Majorana modes in the structure. The model is trained in an unsupervised manner on synthetic data in the form of conductance maps, using a physics-informed loss that incorporates key properties of Majorana zero modes. We show that, with appropriate training, a deep vision-transformer network can efficiently memorize relation between Hamiltonian parameters and structures on conductance maps and use it to propose parameters update for a quantum dot chain that drive the system toward topological phase. Starting from a broad range of initial detunings in parameter space, a single update step is sufficient to generate nontrivial zero modes. Moreover, by enabling an iterative tuning procedure - where the system acquires updated conductance maps at each step - we demonstrate that the method can address a much larger region of the parameter space.
Machine-learned models for magnetic materials
Dec 29, 2023Abstract:We present a general framework for modeling materials using deep neural networks. Material represented by multidimensional characteristics (that mimic measurements) is used to train the neural autoencoder model in an unsupervised manner. The encoder is trying to predict the material parameters of a theoretical model, which is then used in a decoder part. The decoder, using the predicted parameters, reconstructs the input characteristics. The neural model is trained to capture a synthetically generated set of characteristics that can cover a broad range of material behaviors, leading to a model that can generalize on the underlying physics rather than just optimize the model parameters for a single measurement. After setting up the model we prove its usefulness in the complex problem of modeling magnetic materials in the frequency and current (out-of-linear range) domains simultaneously.
Data-driven criteria for quantum correlations
Jul 20, 2023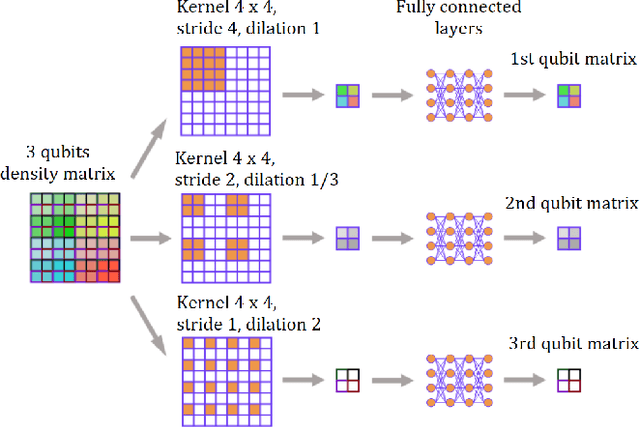
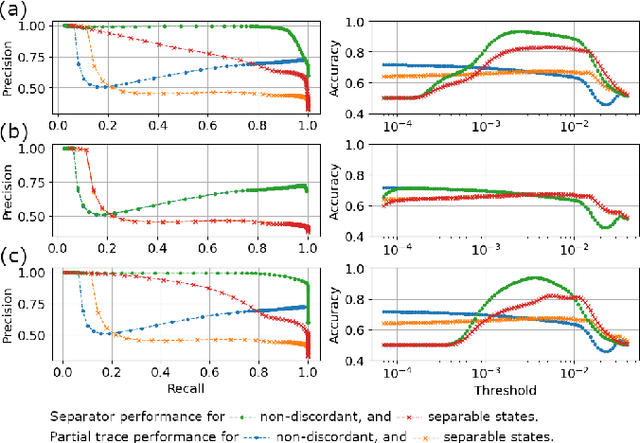
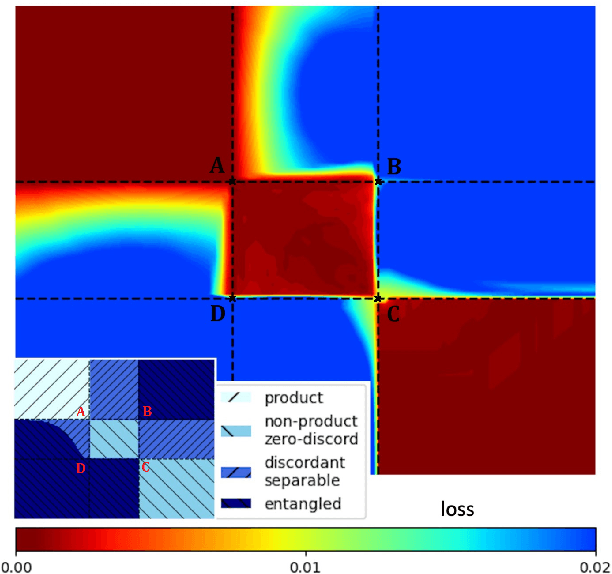
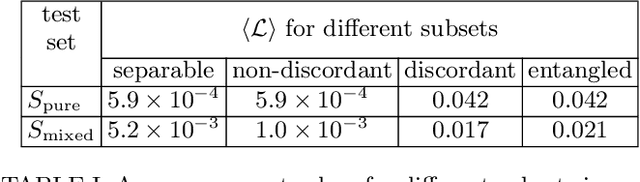
Abstract:We build a machine learning model to detect correlations in a three-qubit system using a neural network trained in an unsupervised manner on randomly generated states. The network is forced to recognize separable states, and correlated states are detected as anomalies. Quite surprisingly, we find that the proposed detector performs much better at distinguishing a weaker form of quantum correlations, namely, the quantum discord, than entanglement. In fact, it has a tendency to grossly overestimate the set of entangled states even at the optimal threshold for entanglement detection, while it underestimates the set of discordant states to a much lesser extent. In order to illustrate the nature of states classified as quantum-correlated, we construct a diagram containing various types of states -- entangled, as well as separable, both discordant and non-discordant. We find that the near-zero value of the recognition loss reproduces the shape of the non-discordant separable states with high accuracy, especially considering the non-trivial shape of this set on the diagram. The network architecture is designed carefully: it preserves separability, and its output is equivariant with respect to qubit permutations. We show that the choice of architecture is important to get the highest detection accuracy, much better than for a baseline model that just utilizes a partial trace operation.
Quantification of entanglement with Siamese convolutional neural networks
Oct 13, 2022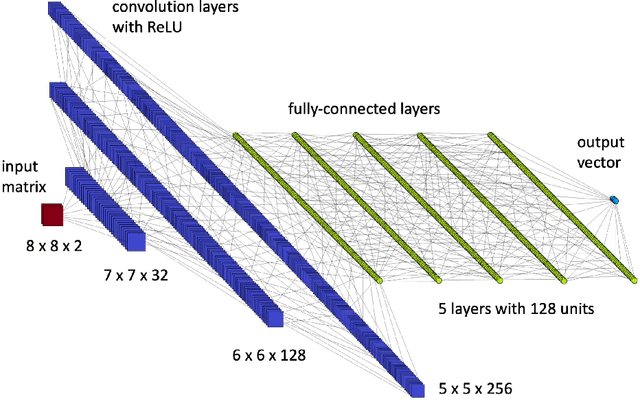
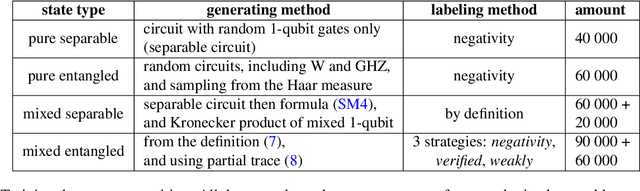
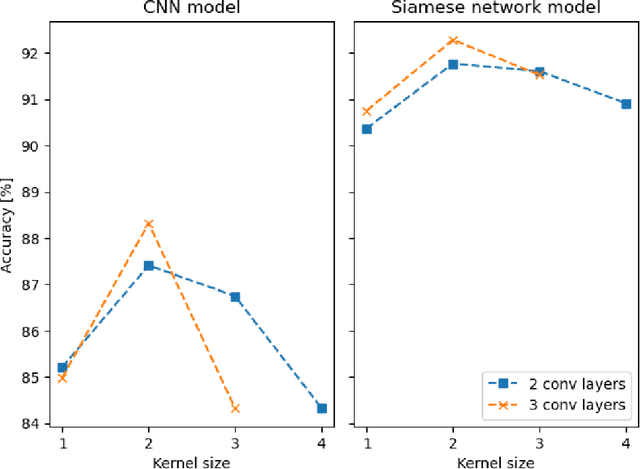
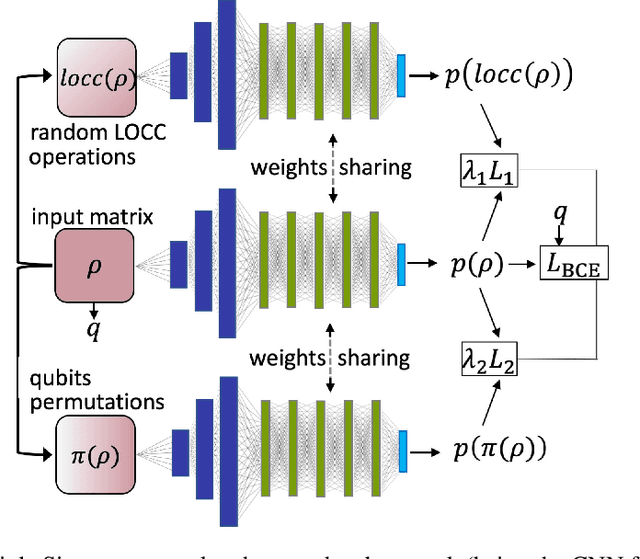
Abstract:Quantum entanglement is a fundamental property commonly used in various quantum information protocols and algorithms. Nonetheless, the problem of quantifying entanglement has still not reached general solution for systems larger than two qubits. In this paper, we investigate the possibility of detecting entanglement with the use of the supervised machine learning method, namely the deep convolutional neural networks. We build a model consisting of convolutional layers, which is able to recognize and predict the presence of entanglement for any bipartition of the given multi-qubit system. We demonstrate that training our model on synthetically generated datasets collecting random density matrices, which either include or exclude challenging positive-under-partial-transposition entangled states (PPTES), leads to the different accuracy of the model and its possibility to detect such states. Moreover, it is shown that enforcing entanglement-preserving symmetry operations (local operations on qubit or permutations of qubits) by using triple Siamese network, can significantly increase the model performance and ability to generalize on types of states not seen during the training stage. We perform numerical calculations for 3,4 and 5-qubit systems, therefore proving the scalability of the proposed approach.
Self-Normalized Density Map (SNDM) for Counting Microbiological Objects
Mar 15, 2022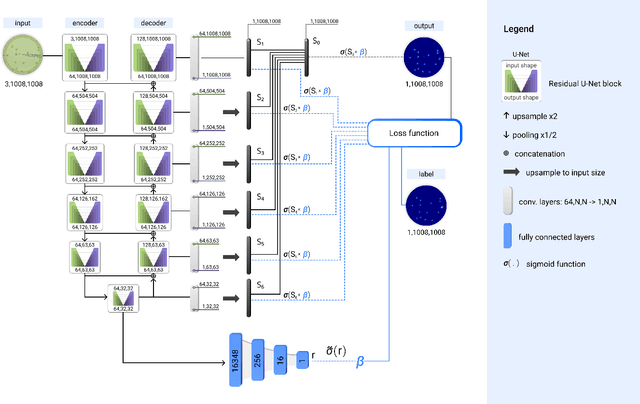
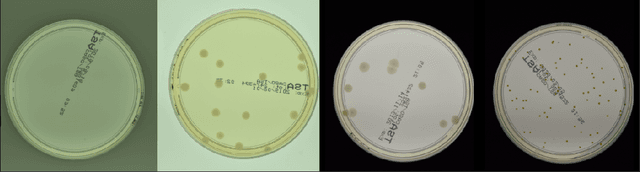
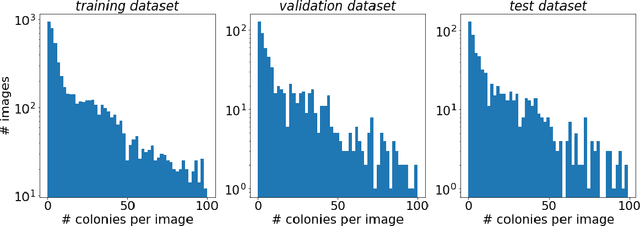
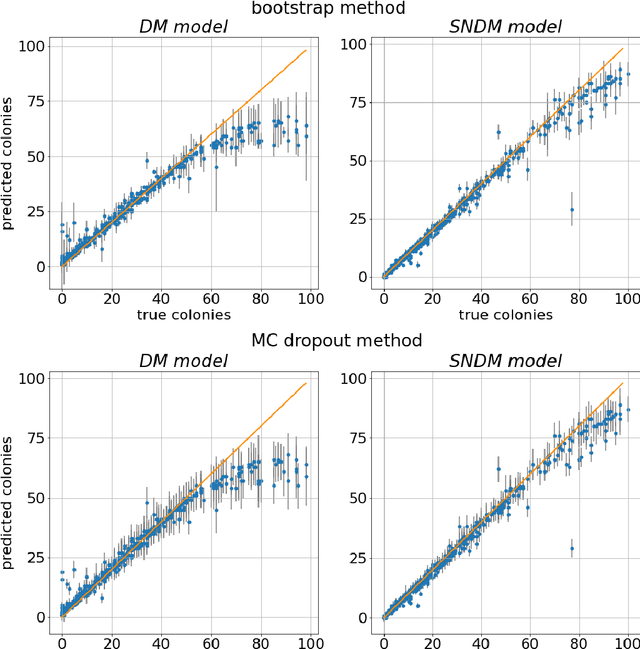
Abstract:The statistical properties of the density map (DM) approach to counting microbiological objects on images are studied in detail. The DM is given by U$^2$-Net. Two statistical methods for deep neural networks are utilized: the bootstrap and the Monte Carlo (MC) dropout. The detailed analysis of the uncertainties for the DM predictions leads to a deeper understanding of the DM model's deficiencies. Based on our investigation, we propose a self-normalization module in the network. The improved network model, called Self-Normalized Density Map (SNDM), can correct its output density map by itself to accurately predict the total number of objects in the image. The SNDM architecture outperforms the original model. Moreover, both statistical frameworks -- bootstrap and MC dropout -- have consistent statistical results for SNDM, which were not observed in the original model.
Generation of microbial colonies dataset with deep learning style transfer
Nov 06, 2021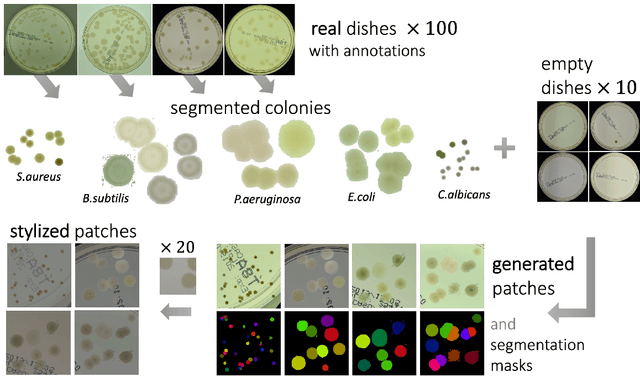
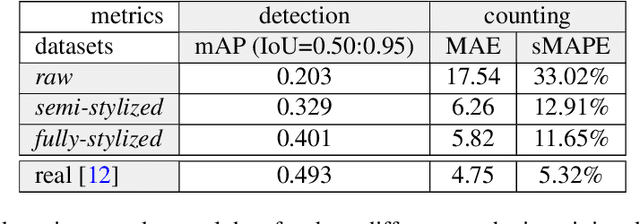
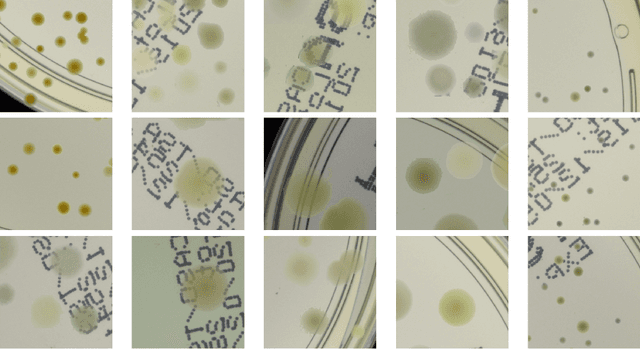
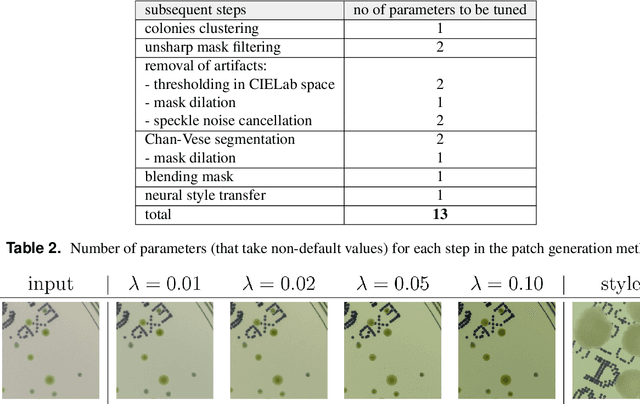
Abstract:We introduce an effective strategy to generate a synthetic dataset of microbiological images of Petri dishes that can be used to train deep learning models. The developed generator employs traditional computer vision algorithms together with a neural style transfer method for data augmentation. We show that the method is able to synthesize a dataset of realistic looking images that can be used to train a neural network model capable of localising, segmenting, and classifying five different microbial species. Our method requires significantly fewer resources to obtain a useful dataset than collecting and labeling a whole large set of real images with annotations. We show that starting with only 100 real images, we can generate data to train a detector that achieves comparable results to the same detector but trained on a real, several dozen times bigger dataset. We prove the usefulness of the method in microbe detection and segmentation, but we expect that it is general and flexible and can also be applicable in other domains of science and industry to detect various objects.
Deep neural networks approach to microbial colony detection -- a comparative analysis
Aug 24, 2021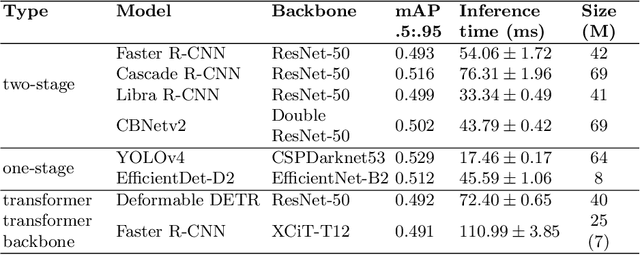
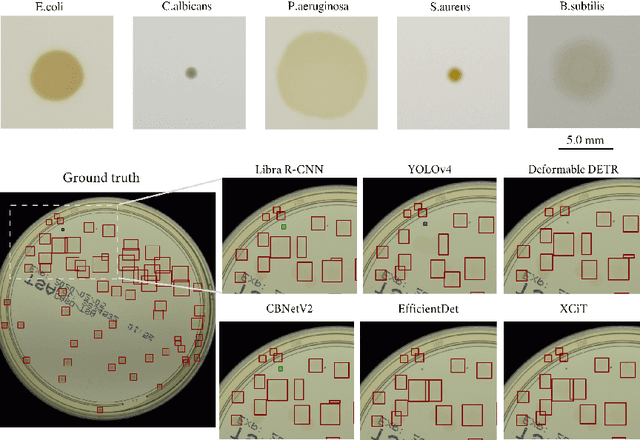
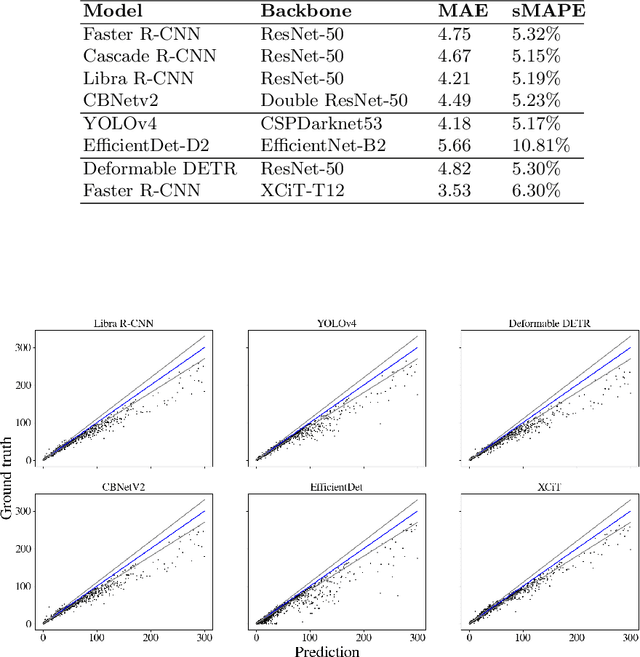
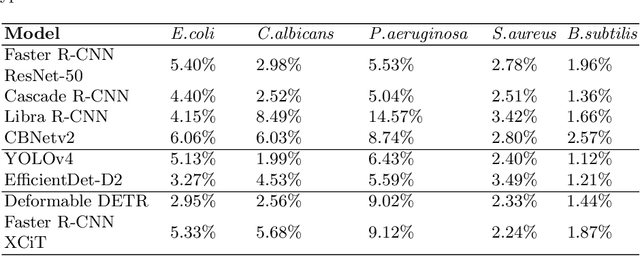
Abstract:Counting microbial colonies is a fundamental task in microbiology and has many applications in numerous industry branches. Despite this, current studies towards automatic microbial counting using artificial intelligence are hardly comparable due to the lack of unified methodology and the availability of large datasets. The recently introduced AGAR dataset is the answer to the second need, but the research carried out is still not exhaustive. To tackle this problem, we compared the performance of three well-known deep learning approaches for object detection on the AGAR dataset, namely two-stage, one-stage and transformer based neural networks. The achieved results may serve as a benchmark for future experiments.
AGAR a microbial colony dataset for deep learning detection
Aug 03, 2021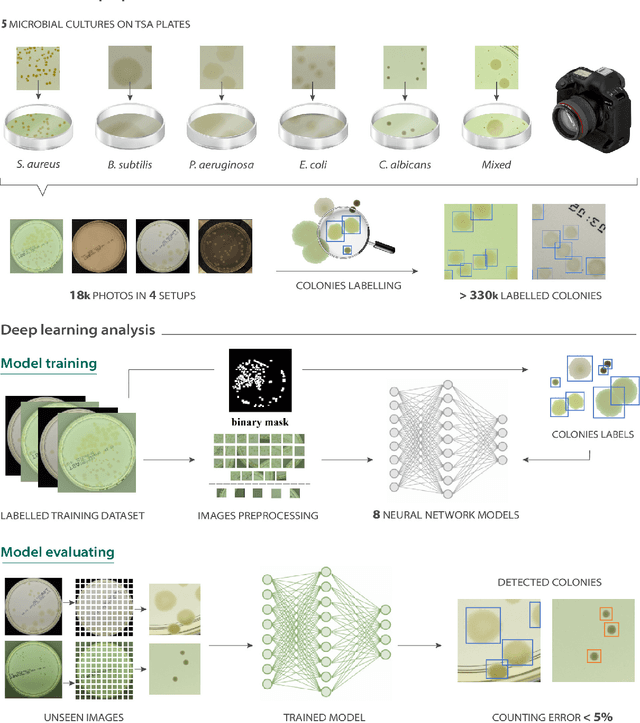
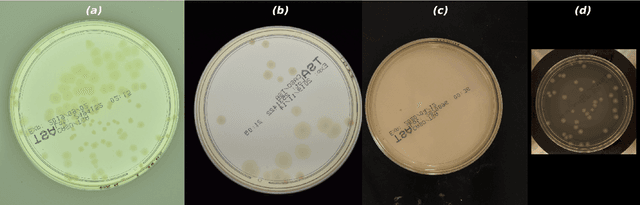
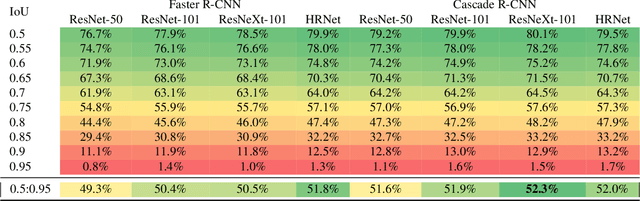
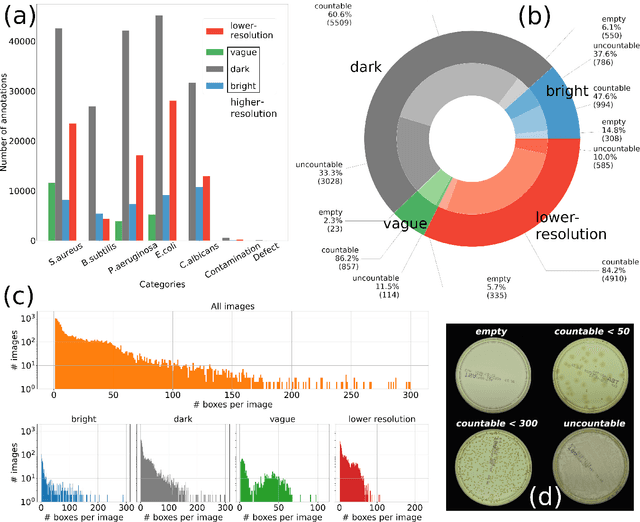
Abstract:The Annotated Germs for Automated Recognition (AGAR) dataset is an image database of microbial colonies cultured on agar plates. It contains 18000 photos of five different microorganisms as single or mixed cultures, taken under diverse lighting conditions with two different cameras. All the images are classified into "countable", "uncountable", and "empty", with the "countable" class labeled by microbiologists with colony location and species identification (336442 colonies in total). This study describes the dataset itself and the process of its development. In the second part, the performance of selected deep neural network architectures for object detection, namely Faster R-CNN and Cascade R-CNN, was evaluated on the AGAR dataset. The results confirmed the great potential of deep learning methods to automate the process of microbe localization and classification based on Petri dish photos. Moreover, AGAR is the first publicly available dataset of this kind and size and will facilitate the future development of machine learning models. The data used in these studies can be found at https://agar.neurosys.com/.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge