Sunil Mohan
How Well Do LLMs Understand Drug Mechanisms? A Knowledge + Reasoning Evaluation Dataset
Nov 09, 2025Abstract:Two scientific fields showing increasing interest in pre-trained large language models (LLMs) are drug development / repurposing, and personalized medicine. For both, LLMs have to demonstrate factual knowledge as well as a deep understanding of drug mechanisms, so they can recall and reason about relevant knowledge in novel situations. Drug mechanisms of action are described as a series of interactions between biomedical entities, which interlink into one or more chains directed from the drug to the targeted disease. Composing the effects of the interactions in a candidate chain leads to an inference about whether the drug might be useful or not for that disease. We introduce a dataset that evaluates LLMs on both factual knowledge of known mechanisms, and their ability to reason about them under novel situations, presented as counterfactuals that the models are unlikely to have seen during training. Using this dataset, we show that o4-mini outperforms the 4o, o3, and o3-mini models from OpenAI, and the recent small Qwen3-4B-thinking model closely matches o4-mini's performance, even outperforming it in some cases. We demonstrate that the open world setting for reasoning tasks, which requires the model to recall relevant knowledge, is more challenging than the closed world setting where the needed factual knowledge is provided. We also show that counterfactuals affecting internal links in the reasoning chain present a much harder task than those affecting a link from the drug mentioned in the prompt.
A Distant Supervision Corpus for Extracting Biomedical Relationships Between Chemicals, Diseases and Genes
Apr 13, 2022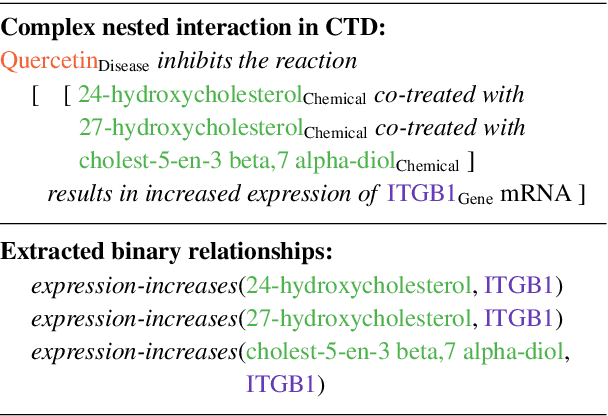
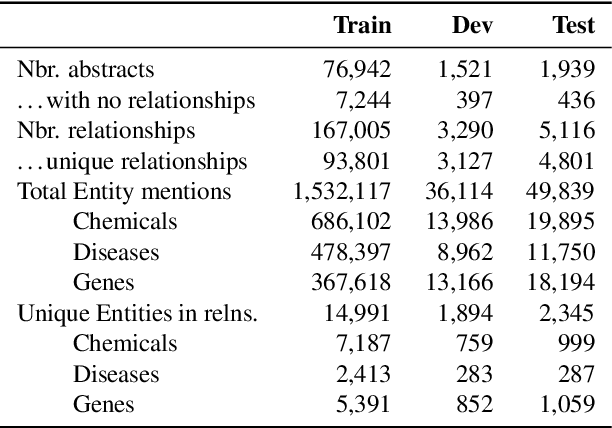
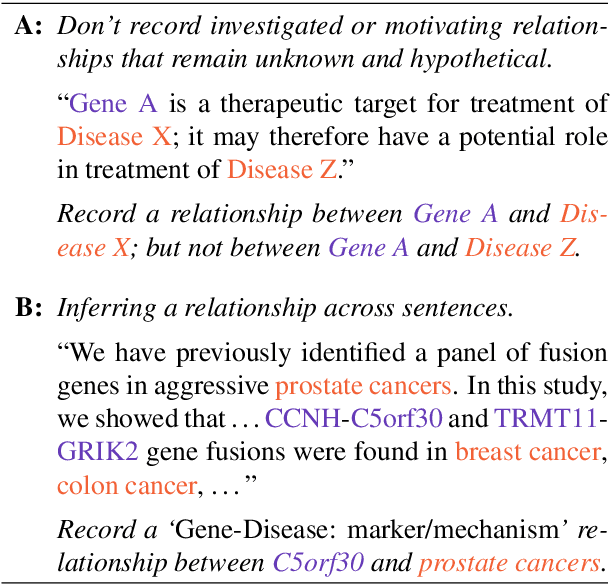
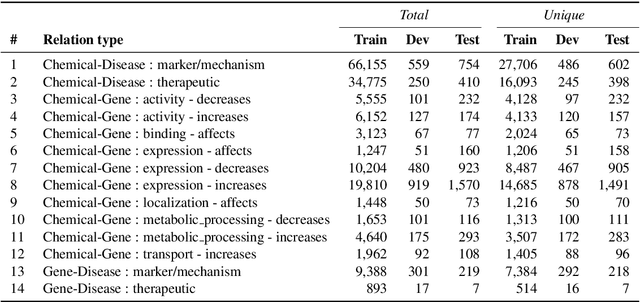
Abstract:We introduce ChemDisGene, a new dataset for training and evaluating multi-class multi-label document-level biomedical relation extraction models. Our dataset contains 80k biomedical research abstracts labeled with mentions of chemicals, diseases, and genes, portions of which human experts labeled with 18 types of biomedical relationships between these entities (intended for evaluation), and the remainder of which (intended for training) has been distantly labeled via the CTD database with approximately 78\% accuracy. In comparison to similar preexisting datasets, ours is both substantially larger and cleaner; it also includes annotations linking mentions to their entities. We also provide three baseline deep neural network relation extraction models trained and evaluated on our new dataset.
LSTM-RASA Based Agri Farm Assistant for Farmers
Apr 07, 2022
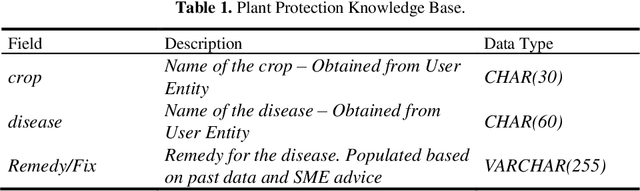


Abstract:The application of Deep Learning and Natural Language based ChatBots are growing rapidly in recent years. They are used in many fields like customer support, reservation system and as personal assistant. The Enterprises are using such ChatBots to serve their customers in a better and efficient manner. Even after such technological advancement, the expert advice does not reach the farmers on timely manner. The farmers are still largely dependent on their peers knowledge in solving the problems they face in their field. These technologies have not been effectively used to give the required information to farmers on timely manner. This project aims to implement a closed domain ChatBot for the field of Agriculture Farmers Assistant. Farmers can have conversation with the Chatbot and get the expert advice in their field. Farmers Assistant is based on RASA Open Source Framework. The Chatbot identifies the intent and entity from user utterances and retrieve the remedy from the database and share it with the user. We tested the Bot with existing data and it showed promising results.
Low Resource Recognition and Linking of Biomedical Concepts from a Large Ontology
Jan 27, 2021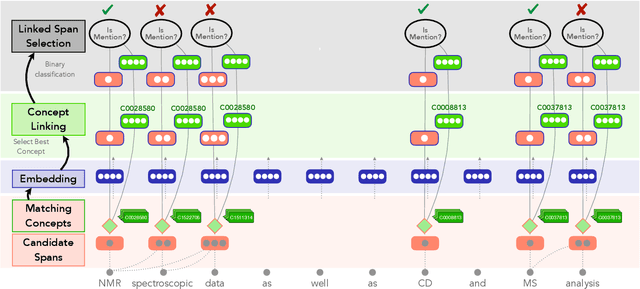
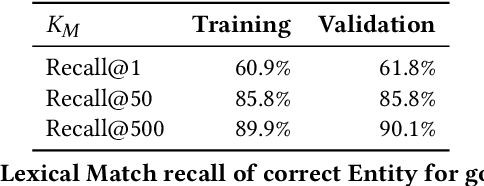
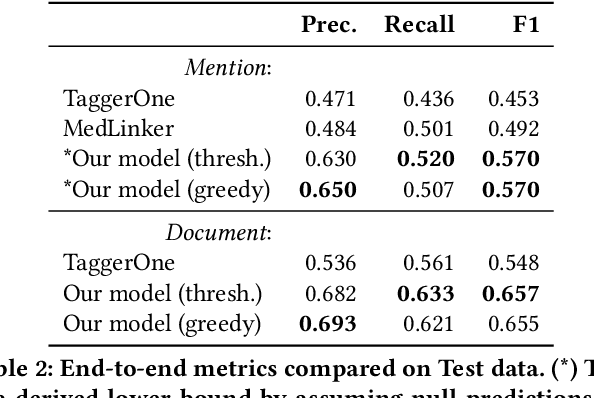
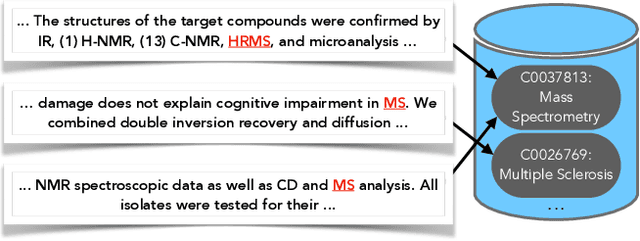
Abstract:Tools to explore scientific literature are essential for scientists, especially in biomedicine, where about a million new papers are published every year. Many such tools provide users the ability to search for specific entities (e.g. proteins, diseases) by tracking their mentions in papers. PubMed, the most well known database of biomedical papers, relies on human curators to add these annotations. This can take several weeks for new papers, and not all papers get tagged. Machine learning models have been developed to facilitate the semantic indexing of scientific papers. However their performance on the more comprehensive ontologies of biomedical concepts does not reach the levels of typical entity recognition problems studied in NLP. In large part this is due to their low resources, where the ontologies are large, there is a lack of descriptive text defining most entities, and labeled data can only cover a small portion of the ontology. In this paper, we develop a new model that overcomes these challenges by (1) generalizing to entities unseen at training time, and (2) incorporating linking predictions into the mention segmentation decisions. Our approach achieves new state-of-the-art results for the UMLS ontology in both traditional recognition/linking (+8 F1 pts) as well as semantic indexing-based evaluation (+10 F1 pts).
Clustering-based Inference for Zero-Shot Biomedical Entity Linking
Oct 21, 2020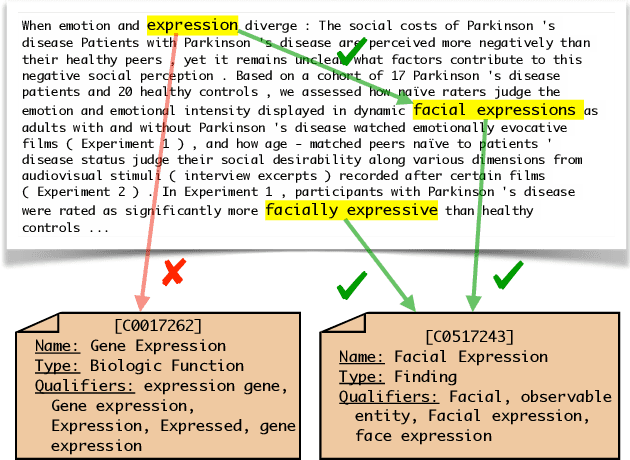
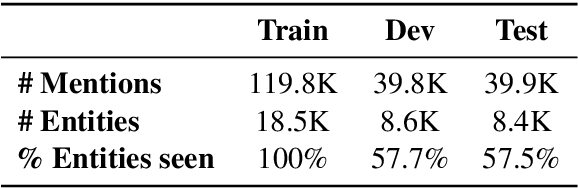
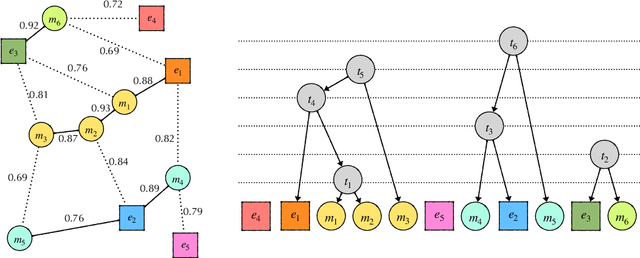
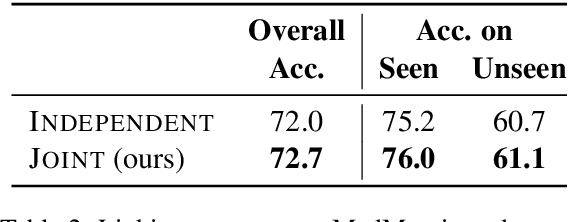
Abstract:Due to large number of entities in biomedical knowledge bases, only a small fraction of entities have corresponding labelled training data. This necessitates a zero-shot entity linking model which is able to link mentions of unseen entities using learned representations of entities. Existing zero-shot entity linking models however link each mention independently, ignoring the inter/intra-document relationships between the entity mentions. These relations can be very useful for linking mentions in biomedical text where linking decisions are often difficult due mentions having a generic or a highly specialized form. In this paper, we introduce a model in which linking decisions can be made not merely by linking to a KB entity but also by grouping multiple mentions together via clustering and jointly making linking predictions. In experiments on the largest publicly available biomedical dataset, we improve the best independent prediction for zero-shot entity linking by 2.5 points of accuracy, and our joint inference model further improves entity linking by 1.8 points.
Overcoming Practical Issues of Deep Active Learning and its Applications on Named Entity Recognition
Nov 17, 2019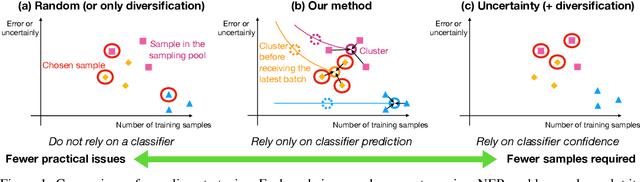
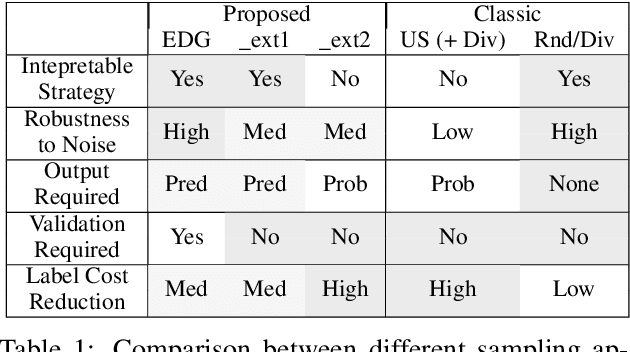
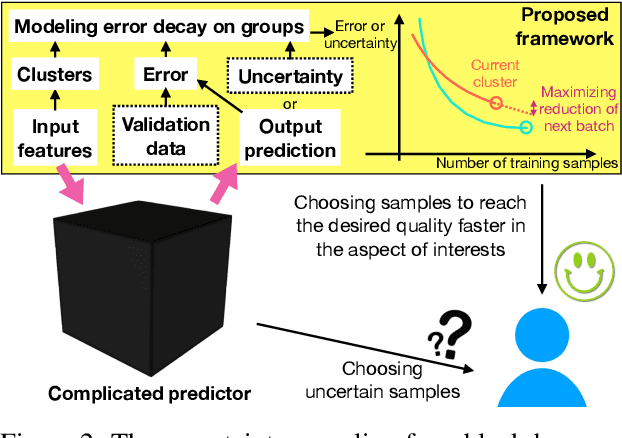
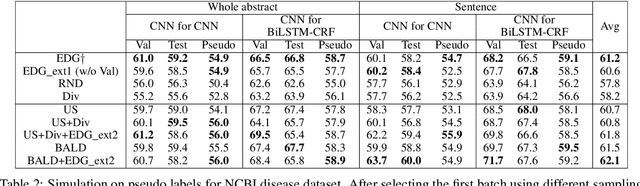
Abstract:Existing deep active learning algorithms achieve impressive sampling efficiency on natural language processing tasks. However, they exhibit several weaknesses in practice, including (a) inability to use uncertainty sampling with black-box models, (b) lack of robustness to noise in labeling, (c) lack of transparency. In response, we propose a transparent batch active sampling framework by estimating the error decay curves of multiple feature-defined subsets of the data. Experiments on four named entity recognition (NER) tasks demonstrate that the proposed methods significantly outperform diversification-based methods for black-box NER taggers and can make the sampling process more robust to labeling noise when combined with uncertainty-based methods. Furthermore, the analysis of experimental results sheds light on the weaknesses of different active sampling strategies, and when traditional uncertainty-based or diversification-based methods can be expected to work well.
MedMentions: A Large Biomedical Corpus Annotated with UMLS Concepts
Feb 25, 2019
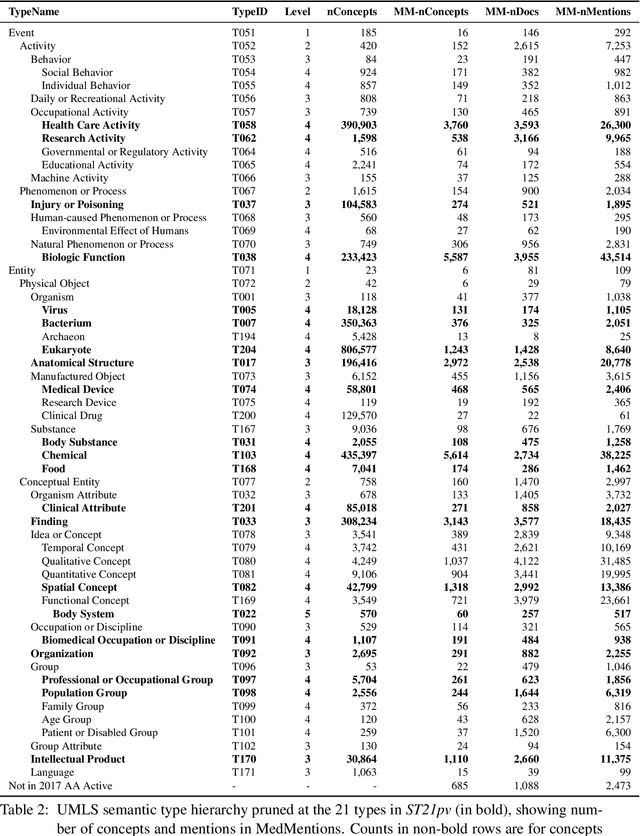
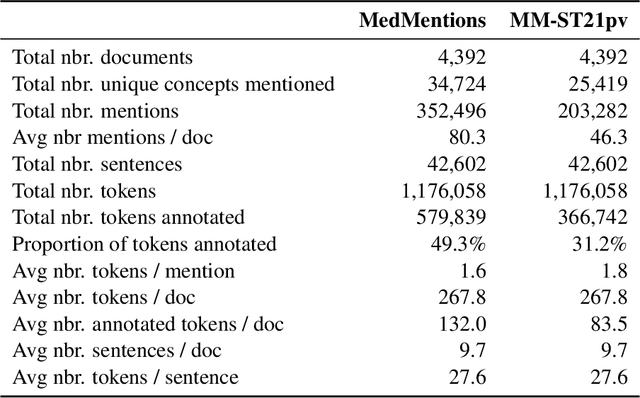

Abstract:This paper presents the formal release of MedMentions, a new manually annotated resource for the recognition of biomedical concepts. What distinguishes MedMentions from other annotated biomedical corpora is its size (over 4,000 abstracts and over 350,000 linked mentions), as well as the size of the concept ontology (over 3 million concepts from UMLS 2017) and its broad coverage of biomedical disciplines. In addition to the full corpus, a sub-corpus of MedMentions is also presented, comprising annotations for a subset of UMLS 2017 targeted towards document retrieval. To encourage research in Biomedical Named Entity Recognition and Linking, data splits for training and testing are included in the release, and a baseline model and its metrics for entity linking are also described.
A Fast Deep Learning Model for Textual Relevance in Biomedical Information Retrieval
Feb 26, 2018


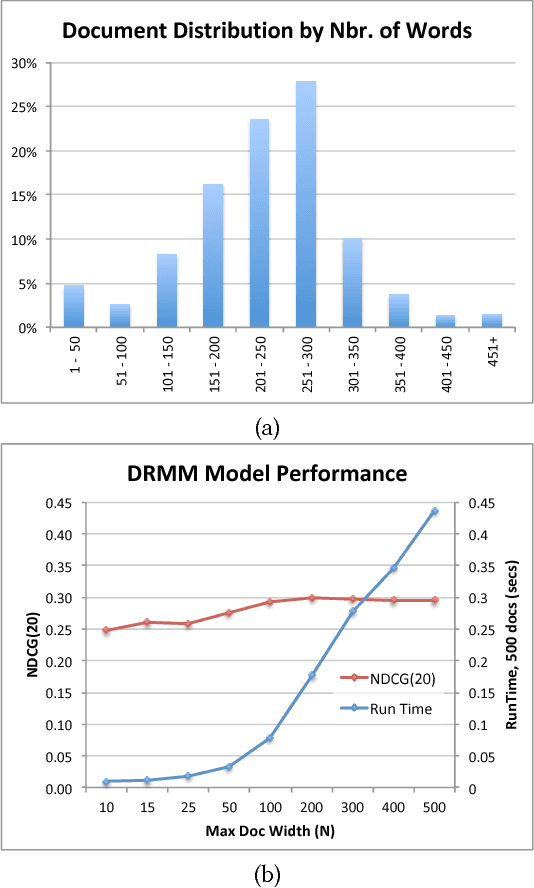
Abstract:Publications in the life sciences are characterized by a large technical vocabulary, with many lexical and semantic variations for expressing the same concept. Towards addressing the problem of relevance in biomedical literature search, we introduce a deep learning model for the relevance of a document's text to a keyword style query. Limited by a relatively small amount of training data, the model uses pre-trained word embeddings. With these, the model first computes a variable-length Delta matrix between the query and document, representing a difference between the two texts, which is then passed through a deep convolution stage followed by a deep feed-forward network to compute a relevance score. This results in a fast model suitable for use in an online search engine. The model is robust and outperforms comparable state-of-the-art deep learning approaches.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge