Clustering-based Inference for Zero-Shot Biomedical Entity Linking
Paper and Code
Oct 21, 2020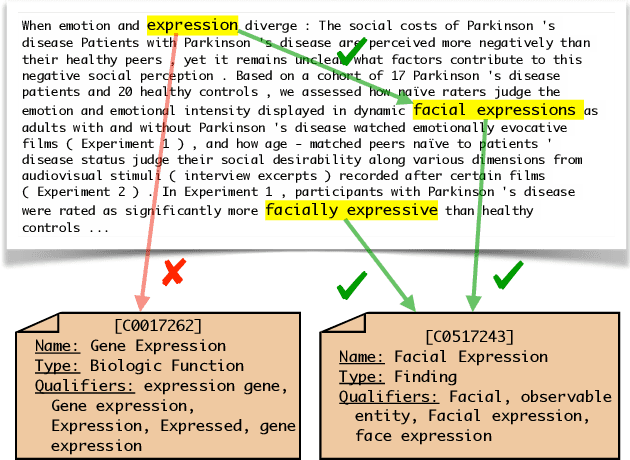
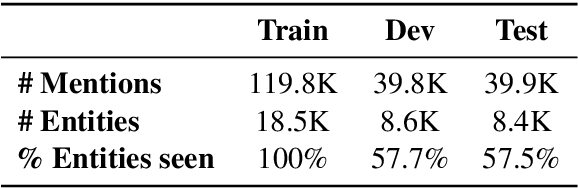
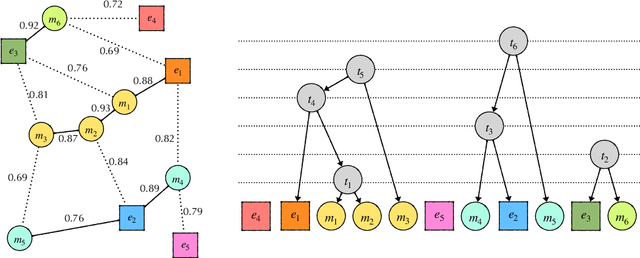
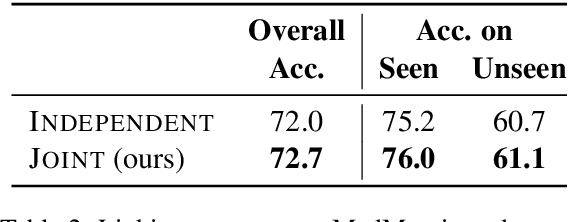
Due to large number of entities in biomedical knowledge bases, only a small fraction of entities have corresponding labelled training data. This necessitates a zero-shot entity linking model which is able to link mentions of unseen entities using learned representations of entities. Existing zero-shot entity linking models however link each mention independently, ignoring the inter/intra-document relationships between the entity mentions. These relations can be very useful for linking mentions in biomedical text where linking decisions are often difficult due mentions having a generic or a highly specialized form. In this paper, we introduce a model in which linking decisions can be made not merely by linking to a KB entity but also by grouping multiple mentions together via clustering and jointly making linking predictions. In experiments on the largest publicly available biomedical dataset, we improve the best independent prediction for zero-shot entity linking by 2.5 points of accuracy, and our joint inference model further improves entity linking by 1.8 points.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge