Sohan Seth
TraCeR: Transformer-Based Competing Risk Analysis with Longitudinal Covariates
Dec 19, 2025



Abstract:Survival analysis is a critical tool for modeling time-to-event data. Recent deep learning-based models have reduced various modeling assumptions including proportional hazard and linearity. However, a persistent challenge remains in incorporating longitudinal covariates, with prior work largely focusing on cross-sectional features, and in assessing calibration of these models, with research primarily focusing on discrimination during evaluation. We introduce TraCeR, a transformer-based survival analysis framework for incorporating longitudinal covariates. Based on a factorized self-attention architecture, TraCeR estimates the hazard function from a sequence of measurements, naturally capturing temporal covariate interactions without assumptions about the underlying data-generating process. The framework is inherently designed to handle censored data and competing events. Experiments on multiple real-world datasets demonstrate that TraCeR achieves substantial and statistically significant performance improvements over state-of-the-art methods. Furthermore, our evaluation extends beyond discrimination metrics and assesses model calibration, addressing a key oversight in literature.
Census-Independent Population Estimation using Representation Learning
Oct 06, 2021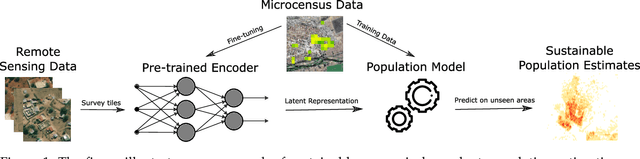
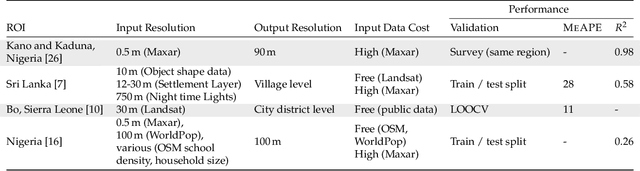
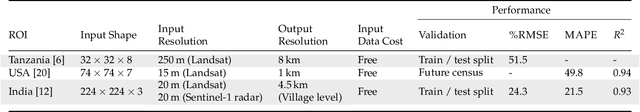
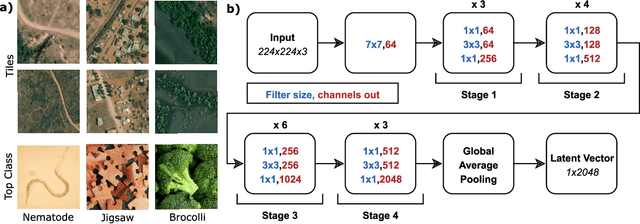
Abstract:Knowledge of population distribution is critical for building infrastructure, distributing resources, and monitoring the progress of sustainable development goals. Although censuses can provide this information, they are typically conducted every ten years with some countries having forgone the process for several decades. Population can change in the intercensal period due to rapid migration, development, urbanisation, natural disasters, and conflicts. Census-independent population estimation approaches using alternative data sources, such as satellite imagery, have shown promise in providing frequent and reliable population estimates locally. Existing approaches, however, require significant human supervision, for example annotating buildings and accessing various public datasets, and therefore, are not easily reproducible. We explore recent representation learning approaches, and assess the transferability of representations to population estimation in Mozambique. Using representation learning reduces required human supervision, since features are extracted automatically, making the process of population estimation more sustainable and likely to be transferable to other regions or countries. We compare the resulting population estimates to existing population products from GRID3, Facebook (HRSL) and WorldPop. We observe that our approach matches the most accurate of these maps, and is interpretable in the sense that it recognises built-up areas to be an informative indicator of population.
Towards Sustainable Census Independent Population Estimation in Mozambique
Apr 26, 2021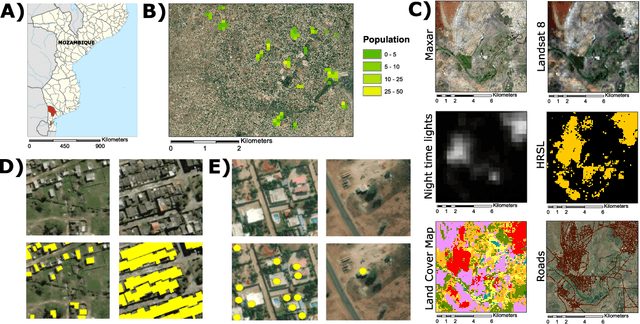
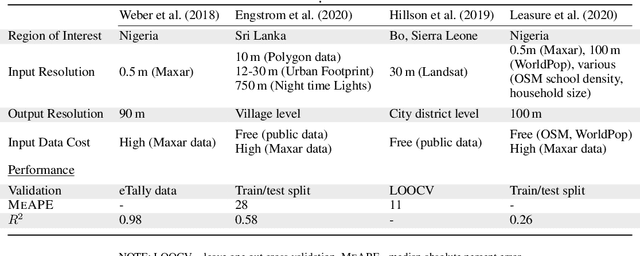
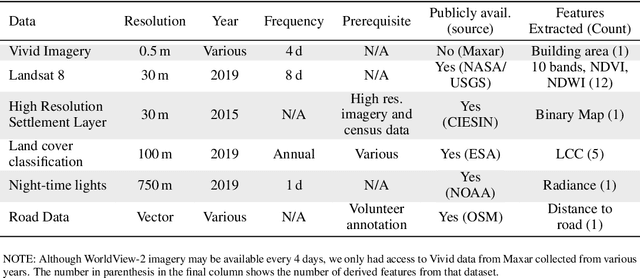
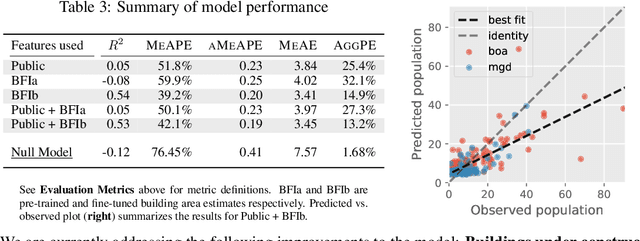
Abstract:Reliable and frequent population estimation is key for making policies around vaccination and planning infrastructure delivery. Since censuses lack the spatio-temporal resolution required for these tasks, census-independent approaches, using remote sensing and microcensus data, have become popular. We estimate intercensal population count in two pilot districts in Mozambique. To encourage sustainability, we assess the feasibility of using publicly available datasets to estimate population. We also explore transfer learning with existing annotated datasets for predicting building footprints, and training with additional `dot' annotations from regions of interest to enhance these estimations. We observe that population predictions improve when using footprint area estimated with this approach versus only publicly available features.
Model Criticism in Latent Space
Jul 02, 2018



Abstract:Model criticism is usually carried out by assessing if replicated data generated under the fitted model looks similar to the observed data, see e.g. Gelman, Carlin, Stern, and Rubin [2004, p. 165]. This paper presents a method for latent variable models by pulling back the data into the space of latent variables, and carrying out model criticism in that space. Making use of a model's structure enables a more direct assessment of the assumptions made in the prior and likelihood. We demonstrate the method with examples of model criticism in latent space applied to factor analysis, linear dynamical systems and Gaussian processes.
Modelling-based experiment retrieval: A case study with gene expression clustering
Jan 04, 2016



Abstract:Motivation: Public and private repositories of experimental data are growing to sizes that require dedicated methods for finding relevant data. To improve on the state of the art of keyword searches from annotations, methods for content-based retrieval have been proposed. In the context of gene expression experiments, most methods retrieve gene expression profiles, requiring each experiment to be expressed as a single profile, typically of case vs. control. A more general, recently suggested alternative is to retrieve experiments whose models are good for modelling the query dataset. However, for very noisy and high-dimensional query data, this retrieval criterion turns out to be very noisy as well. Results: We propose doing retrieval using a denoised model of the query dataset, instead of the original noisy dataset itself. To this end, we introduce a general probabilistic framework, where each experiment is modelled separately and the retrieval is done by finding related models. For retrieval of gene expression experiments, we use a probabilistic model called product partition model, which induces a clustering of genes that show similar expression patterns across a number of samples. The suggested metric for retrieval using clusterings is the normalized information distance. Empirical results finally suggest that inference for the full probabilistic model can be approximated with good performance using computationally faster heuristic clustering approaches (e.g. $k$-means). The method is highly scalable and straightforward to apply to construct a general-purpose gene expression experiment retrieval method. Availability: The method can be implemented using standard clustering algorithms and normalized information distance, available in many statistical software packages.
Probabilistic Archetypal Analysis
Apr 07, 2014



Abstract:Archetypal analysis represents a set of observations as convex combinations of pure patterns, or archetypes. The original geometric formulation of finding archetypes by approximating the convex hull of the observations assumes them to be real valued. This, unfortunately, is not compatible with many practical situations. In this paper we revisit archetypal analysis from the basic principles, and propose a probabilistic framework that accommodates other observation types such as integers, binary, and probability vectors. We corroborate the proposed methodology with convincing real-world applications on finding archetypal winter tourists based on binary survey data, archetypal disaster-affected countries based on disaster count data, and document archetypes based on term-frequency data. We also present an appropriate visualization tool to summarize archetypal analysis solution better.
Retrieval of Experiments with Sequential Dirichlet Process Mixtures in Model Space
Mar 06, 2014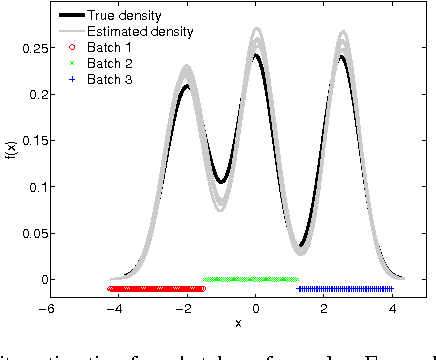
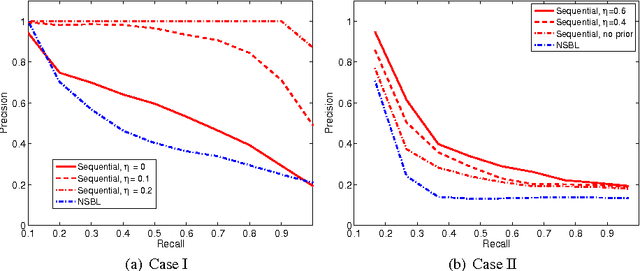
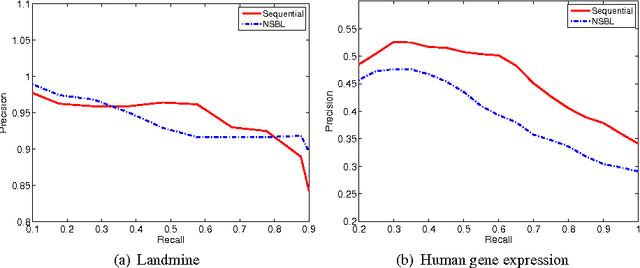
Abstract:We address the problem of retrieving relevant experiments given a query experiment, motivated by the public databases of datasets in molecular biology and other experimental sciences, and the need of scientists to relate to earlier work on the level of actual measurement data. Since experiments are inherently noisy and databases ever accumulating, we argue that a retrieval engine should possess two particular characteristics. First, it should compare models learnt from the experiments rather than the raw measurements themselves: this allows incorporating experiment-specific prior knowledge to suppress noise effects and focus on what is important. Second, it should be updated sequentially from newly published experiments, without explicitly storing either the measurements or the models, which is critical for saving storage space and protecting data privacy: this promotes life long learning. We formulate the retrieval as a ``supermodelling'' problem, of sequentially learning a model of the set of posterior distributions, represented as sets of MCMC samples, and suggest the use of Particle-Learning-based sequential Dirichlet process mixture (DPM) for this purpose. The relevance measure for retrieval is derived from the supermodel through the mixture representation. We demonstrate the performance of the proposed retrieval method on simulated data and molecular biological experiments.
Retrieval of Experiments by Efficient Estimation of Marginal Likelihood
Feb 19, 2014



Abstract:We study the task of retrieving relevant experiments given a query experiment. By experiment, we mean a collection of measurements from a set of `covariates' and the associated `outcomes'. While similar experiments can be retrieved by comparing available `annotations', this approach ignores the valuable information available in the measurements themselves. To incorporate this information in the retrieval task, we suggest employing a retrieval metric that utilizes probabilistic models learned from the measurements. We argue that such a metric is a sensible measure of similarity between two experiments since it permits inclusion of experiment-specific prior knowledge. However, accurate models are often not analytical, and one must resort to storing posterior samples which demands considerable resources. Therefore, we study strategies to select informative posterior samples to reduce the computational load while maintaining the retrieval performance. We demonstrate the efficacy of our approach on simulated data with simple linear regression as the models, and real world datasets.
Bayesian Extensions of Kernel Least Mean Squares
Oct 20, 2013
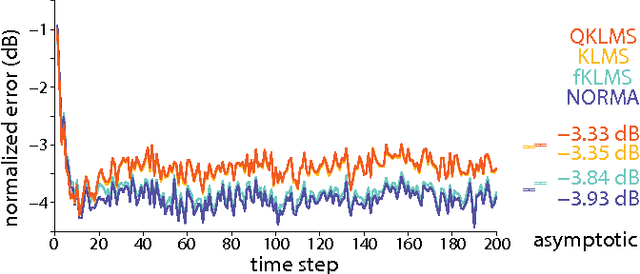
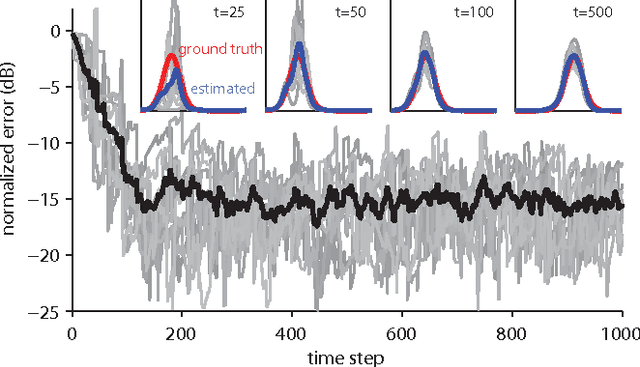

Abstract:The kernel least mean squares (KLMS) algorithm is a computationally efficient nonlinear adaptive filtering method that "kernelizes" the celebrated (linear) least mean squares algorithm. We demonstrate that the least mean squares algorithm is closely related to the Kalman filtering, and thus, the KLMS can be interpreted as an approximate Bayesian filtering method. This allows us to systematically develop extensions of the KLMS by modifying the underlying state-space and observation models. The resulting extensions introduce many desirable properties such as "forgetting", and the ability to learn from discrete data, while retaining the computational simplicity and time complexity of the original algorithm.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge