Sindhu Tipirneni
Context-Aware Clustering using Large Language Models
May 02, 2024Abstract:Despite the remarkable success of Large Language Models (LLMs) in text understanding and generation, their potential for text clustering tasks remains underexplored. We observed that powerful closed-source LLMs provide good quality clusterings of entity sets but are not scalable due to the massive compute power required and the associated costs. Thus, we propose CACTUS (Context-Aware ClusTering with aUgmented triplet losS), a systematic approach that leverages open-source LLMs for efficient and effective supervised clustering of entity subsets, particularly focusing on text-based entities. Existing text clustering methods fail to effectively capture the context provided by the entity subset. Moreover, though there are several language modeling based approaches for clustering, very few are designed for the task of supervised clustering. This paper introduces a novel approach towards clustering entity subsets using LLMs by capturing context via a scalable inter-entity attention mechanism. We propose a novel augmented triplet loss function tailored for supervised clustering, which addresses the inherent challenges of directly applying the triplet loss to this problem. Furthermore, we introduce a self-supervised clustering task based on text augmentation techniques to improve the generalization of our model. For evaluation, we collect ground truth clusterings from a closed-source LLM and transfer this knowledge to an open-source LLM under the supervised clustering framework, allowing a faster and cheaper open-source model to perform the same task. Experiments on various e-commerce query and product clustering datasets demonstrate that our proposed approach significantly outperforms existing unsupervised and supervised baselines under various external clustering evaluation metrics.
Identifying TBI Physiological States by Clustering of Multivariate Clinical Time-Series
Mar 30, 2023



Abstract:Determining clinically relevant physiological states from multivariate time series data with missing values is essential for providing appropriate treatment for acute conditions such as Traumatic Brain Injury (TBI), respiratory failure, and heart failure. Utilizing non-temporal clustering or data imputation and aggregation techniques may lead to loss of valuable information and biased analyses. In our study, we apply the SLAC-Time algorithm, an innovative self-supervision-based approach that maintains data integrity by avoiding imputation or aggregation, offering a more useful representation of acute patient states. By using SLAC-Time to cluster data in a large research dataset, we identified three distinct TBI physiological states and their specific feature profiles. We employed various clustering evaluation metrics and incorporated input from a clinical domain expert to validate and interpret the identified physiological states. Further, we discovered how specific clinical events and interventions can influence patient states and state transitions.
A Self-Supervised Learning-based Approach to Clustering Multivariate Time-Series Data with Missing Values : An Application to Traumatic Brain Injury Phenotyping
Feb 27, 2023



Abstract:Self-supervised learning approaches provide a promising direction for clustering multivariate time-series data. However, real-world time-series data often include missing values, and the existing approaches require imputing missing values before clustering, which may cause extensive computations and noise and result in invalid interpretations. To address these challenges, we present a Self-supervised Learning-based Approach to Clustering multivariate Time-series data with missing values (SLAC-Time). SLAC-Time is a Transformer-based clustering method that uses time-series forecasting as a proxy task for leveraging unlabeled data and learning more robust time-series representations. This method jointly learns the neural network parameters and the cluster assignments of the learned representations. It iteratively clusters the learned representations with the K-means method and then utilizes the subsequent cluster assignments as pseudo-labels to update the model parameters. To evaluate our proposed approach, we applied it to clustering and phenotyping Traumatic Brain Injury (TBI) patients in the TRACK-TBI dataset. Our experiments demonstrate that SLAC-Time outperforms the baseline K-means clustering algorithm in terms of silhouette coefficient, Calinski Harabasz index, Dunn index, and Davies Bouldin index. We identified three TBI phenotypes that are distinct from one another in terms of clinically significant variables as well as clinical outcomes, including the Extended Glasgow Outcome Scale (GOSE) score, Intensive Care Unit (ICU) length of stay, and mortality rate. The experiments show that the TBI phenotypes identified by SLAC-Time can be potentially used for developing targeted clinical trials and therapeutic strategies.
Execution-based Code Generation using Deep Reinforcement Learning
Feb 13, 2023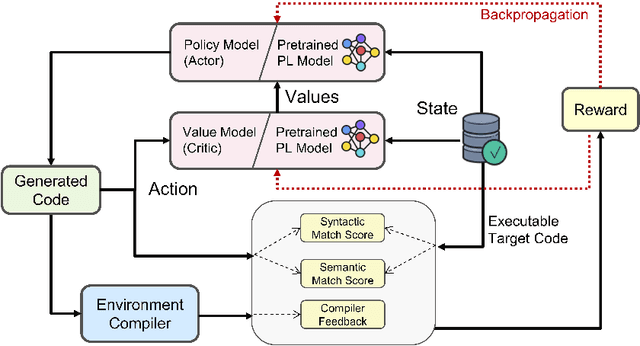
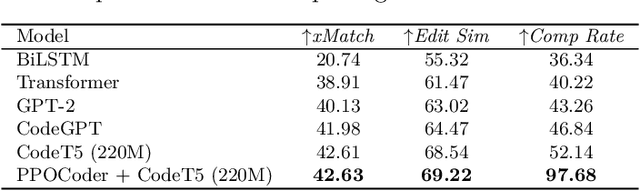
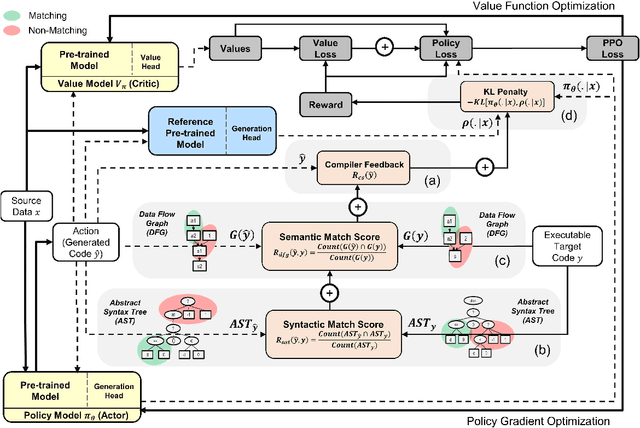
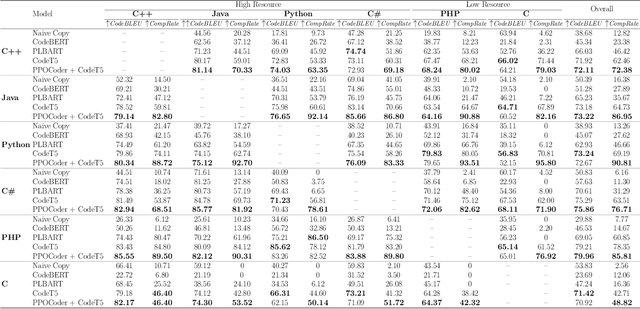
Abstract:The utilization of programming language (PL) models, pretrained on large-scale code corpora, as a means of automating software engineering processes has demonstrated considerable potential in streamlining various code generation tasks such as code completion, code translation, and program synthesis. However, current approaches mainly rely on supervised fine-tuning objectives borrowed from text generation, neglecting specific sequence-level features of code, including but not limited to compilability as well as syntactic and functional correctness. To address this limitation, we propose PPOCoder, a new framework for code generation that combines pretrained PL models with Proximal Policy Optimization (PPO) deep reinforcement learning and employs execution feedback as the external source of knowledge into the model optimization. PPOCoder is transferable across different code generation tasks and PLs. Extensive experiments on three code generation tasks demonstrate the effectiveness of our proposed approach compared to SOTA methods, improving the success rate of compilation and functional correctness over different PLs. Our code can be found at https://github.com/reddy-lab-code-research/PPOCoder .
WindowSHAP: An Efficient Framework for Explaining Time-series Classifiers based on Shapley Values
Nov 11, 2022Abstract:Unpacking and comprehending how deep learning algorithms make decisions has been a persistent challenge for researchers and end-users. Explaining time-series predictive models is useful for clinical applications with high stakes to understand the behavior of prediction models. However, existing approaches to explain such models are frequently unique to architectures and data where the features do not have a time-varying component. In this paper, we introduce WindowSHAP, a model-agnostic framework for explaining time-series classifiers using Shapley values. We intend for WindowSHAP to mitigate the computational complexity of calculating Shapley values for long time-series data as well as improve the quality of explanations. WindowSHAP is based on partitioning a sequence into time windows. Under this framework, we present three distinct algorithms of Stationary, Sliding and Dynamic WindowSHAP, each evaluated against baseline approaches, KernelSHAP and TimeSHAP, using perturbation and sequence analyses metrics. We applied our framework to clinical time-series data from both a specialized clinical domain (Traumatic Brain Injury - TBI) as well as a broad clinical domain (critical care medicine). The experimental results demonstrate that, based on the two quantitative metrics, our framework is superior at explaining clinical time-series classifiers, while also reducing the complexity of computations. We show that for time-series data with 120 time steps (hours), merging 10 adjacent time points can reduce the CPU time of WindowSHAP by 80% compared to KernelSHAP. We also show that our Dynamic WindowSHAP algorithm focuses more on the most important time steps and provides more understandable explanations. As a result, WindowSHAP not only accelerates the calculation of Shapley values for time-series data, but also delivers more understandable explanations with higher quality.
An Empirical Comparison of Explainable Artificial Intelligence Methods for Clinical Data: A Case Study on Traumatic Brain Injury
Aug 13, 2022



Abstract:A longstanding challenge surrounding deep learning algorithms is unpacking and understanding how they make their decisions. Explainable Artificial Intelligence (XAI) offers methods to provide explanations of internal functions of algorithms and reasons behind their decisions in ways that are interpretable and understandable to human users. . Numerous XAI approaches have been developed thus far, and a comparative analysis of these strategies seems necessary to discern their relevance to clinical prediction models. To this end, we first implemented two prediction models for short- and long-term outcomes of traumatic brain injury (TBI) utilizing structured tabular as well as time-series physiologic data, respectively. Six different interpretation techniques were used to describe both prediction models at the local and global levels. We then performed a critical analysis of merits and drawbacks of each strategy, highlighting the implications for researchers who are interested in applying these methodologies. The implemented methods were compared to one another in terms of several XAI characteristics such as understandability, fidelity, and stability. Our findings show that SHAP is the most stable with the highest fidelity but falls short of understandability. Anchors, on the other hand, is the most understandable approach, but it is only applicable to tabular data and not time series data.
XLCoST: A Benchmark Dataset for Cross-lingual Code Intelligence
Jun 16, 2022
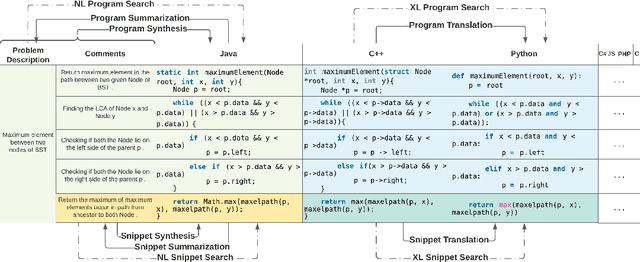
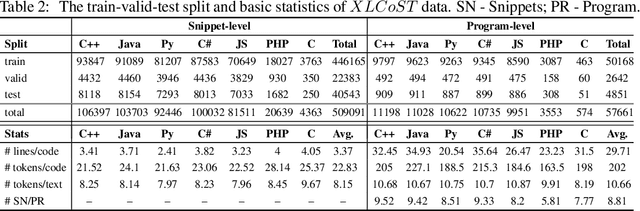
Abstract:Recent advances in machine learning have significantly improved the understanding of source code data and achieved good performance on a number of downstream tasks. Open source repositories like GitHub enable this process with rich unlabeled code data. However, the lack of high quality labeled data has largely hindered the progress of several code related tasks, such as program translation, summarization, synthesis, and code search. This paper introduces XLCoST, Cross-Lingual Code SnippeT dataset, a new benchmark dataset for cross-lingual code intelligence. Our dataset contains fine-grained parallel data from 8 languages (7 commonly used programming languages and English), and supports 10 cross-lingual code tasks. To the best of our knowledge, it is the largest parallel dataset for source code both in terms of size and the number of languages. We also provide the performance of several state-of-the-art baseline models for each task. We believe this new dataset can be a valuable asset for the research community and facilitate the development and validation of new methods for cross-lingual code intelligence.
StructCoder: Structure-Aware Transformer for Code Generation
Jun 10, 2022



Abstract:There has been a recent surge of interest in automating software engineering tasks using deep learning. This work addresses the problem of code generation where the goal is to generate target code given source code in a different language or a natural language description. Most of the state-of-the-art deep learning models for code generation use training strategies that are primarily designed for natural language. However, understanding and generating code requires a more rigorous comprehension of the code syntax and semantics. With this motivation, we develop an encoder-decoder Transformer model where both the encoder and decoder are trained to recognize the syntax and data flow in the source and target codes, respectively. We not only make the encoder structure-aware by leveraging the source code's syntax tree and data flow graph, but we also ensure that our decoder preserves the syntax and data flow of the target code by introducing two auxiliary tasks: AST (Abstract Syntax Tree) paths prediction and data flow prediction. To the best of our knowledge, this is the first work to introduce a structure-aware Transformer decoder to enhance the quality of generated code by modeling target syntax and data flow. The proposed StructCoder model achieves state-of-the-art performance on code translation and text-to-code generation tasks in the CodeXGLUE benchmark.
Self-supervised Transformer for Multivariate Clinical Time-Series with Missing Values
Jul 29, 2021



Abstract:Multivariate time-series (MVTS) data are frequently observed in critical care settings and are typically characterized by excessive missingness and irregular time intervals. Existing approaches for learning representations in this domain handle such issues by either aggregation or imputation of values, which in-turn suppresses the fine-grained information and adds undesirable noise/overhead into the machine learning model. To tackle this challenge, we propose STraTS (Self-supervised Transformer for TimeSeries) model which bypasses these pitfalls by treating time-series as a set of observation triplets instead of using the traditional dense matrix representation. It employs a novel Continuous Value Embedding (CVE) technique to encode continuous time and variable values without the need for discretization. It is composed of a Transformer component with Multi-head attention layers which enables it to learn contextual triplet embeddings while avoiding problems of recurrence and vanishing gradients that occur in recurrent architectures. Many healthcare datasets also suffer from the limited availability of labeled data. Our model utilizes self-supervision by leveraging unlabeled data to learn better representations by performing time-series forecasting as a self-supervision task. Experiments on real-world multivariate clinical time-series benchmark datasets show that STraTS shows better prediction performance than state-of-the-art methods for mortality prediction, especially when labeled data is limited. Finally, we also present an interpretable version of STraTS which can identify important measurements in the time-series data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge