Shuhang Wang
NaviDiffusor: Cost-Guided Diffusion Model for Visual Navigation
Apr 14, 2025Abstract:Visual navigation, a fundamental challenge in mobile robotics, demands versatile policies to handle diverse environments. Classical methods leverage geometric solutions to minimize specific costs, offering adaptability to new scenarios but are prone to system errors due to their multi-modular design and reliance on hand-crafted rules. Learning-based methods, while achieving high planning success rates, face difficulties in generalizing to unseen environments beyond the training data and often require extensive training. To address these limitations, we propose a hybrid approach that combines the strengths of learning-based methods and classical approaches for RGB-only visual navigation. Our method first trains a conditional diffusion model on diverse path-RGB observation pairs. During inference, it integrates the gradients of differentiable scene-specific and task-level costs, guiding the diffusion model to generate valid paths that meet the constraints. This approach alleviates the need for retraining, offering a plug-and-play solution. Extensive experiments in both indoor and outdoor settings, across simulated and real-world scenarios, demonstrate zero-shot transfer capability of our approach, achieving higher success rates and fewer collisions compared to baseline methods. Code will be released at https://github.com/SYSU-RoboticsLab/NaviD.
VEDA: Uneven light image enhancement via a vision-based exploratory data analysis model
May 25, 2023



Abstract:Uneven light image enhancement is a highly demanded task in many industrial image processing applications. Many existing enhancement methods using physical lighting models or deep-learning techniques often lead to unnatural results. This is mainly because: 1) the assumptions and priors made by the physical lighting model (PLM) based approaches are often violated in most natural scenes, and 2) the training datasets or loss functions used by deep-learning technique based methods cannot handle the various lighting scenarios in the real world well. In this paper, we propose a novel vision-based exploratory data analysis model (VEDA) for uneven light image enhancement. Our method is conceptually simple yet effective. A given image is first decomposed into a contrast image that preserves most of the perceptually important scene details, and a residual image that preserves the lighting variations. After achieving this decomposition at multiple scales using a retinal model that simulates the neuron response to light, the enhanced result at each scale can be obtained by manipulating the two images and recombining them. Then, a weighted averaging strategy based on the residual image is designed to obtain the output image by combining enhanced results at multiple scales. A similar weighting strategy can also be leveraged to reconcile noise suppression and detail preservation. Extensive experiments on different image datasets demonstrate that the proposed method can achieve competitive results in its simplicity and effectiveness compared with state-of-the-art methods. It does not require any explicit assumptions and priors about the scene imaging process, nor iteratively solving any optimization functions or any learning procedures.
Brain Tumors Classification for MR images based on Attention Guided Deep Learning Model
Apr 06, 2021
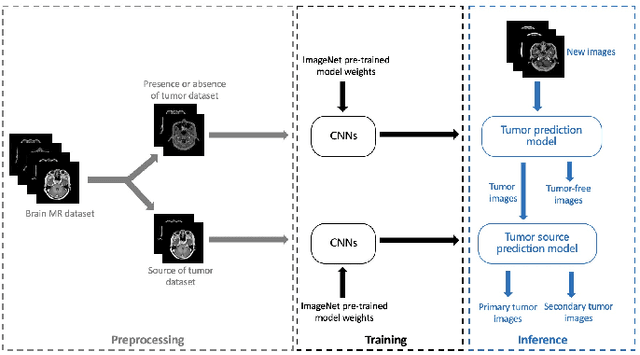
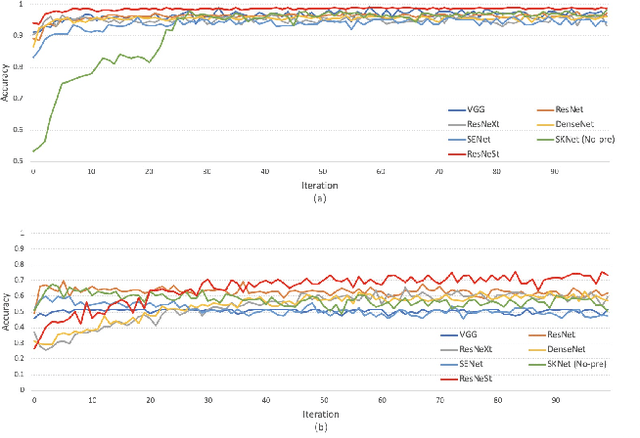
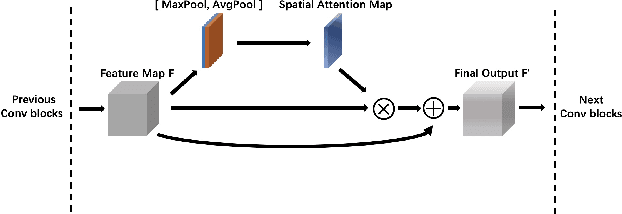
Abstract:In the clinical diagnosis and treatment of brain tumors, manual image reading consumes a lot of energy and time. In recent years, the automatic tumor classification technology based on deep learning has entered people's field of vision. Brain tumors can be divided into primary and secondary intracranial tumors according to their source. However, to our best knowledge, most existing research on brain tumors are limited to primary intracranial tumor images and cannot classify the source of the tumor. In order to solve the task of tumor source type classification, we analyze the existing technology and propose an attention guided deep convolution neural network (CNN) model. Meanwhile, the method proposed in this paper also effectively improves the accuracy of classifying the presence or absence of tumor. For the brain MR dataset, our method can achieve the average accuracy of 99.18% under ten-fold cross-validation for identifying the presence or absence of tumor, and 83.38% for classifying the source of tumor. Experimental results show that our method is consistent with the method of medical experts. It can assist doctors in achieving efficient clinical diagnosis of brain tumors.
Network-Agnostic Knowledge Transfer for Medical Image Segmentation
Jan 23, 2021
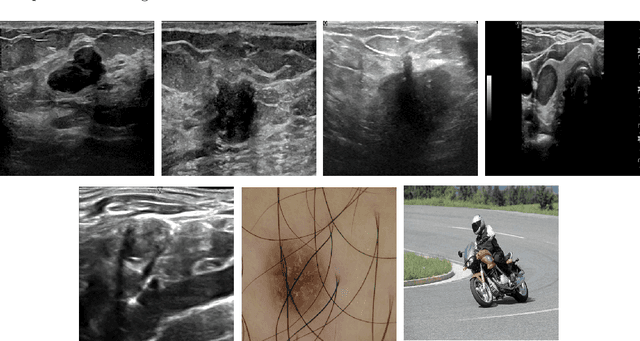
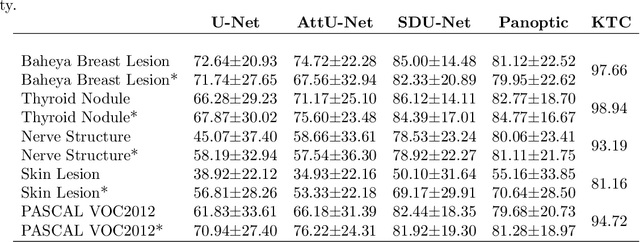
Abstract:Conventional transfer learning leverages weights of pre-trained networks, but mandates the need for similar neural architectures. Alternatively, knowledge distillation can transfer knowledge between heterogeneous networks but often requires access to the original training data or additional generative networks. Knowledge transfer between networks can be improved by being agnostic to the choice of network architecture and reducing the dependence on original training data. We propose a knowledge transfer approach from a teacher to a student network wherein we train the student on an independent transferal dataset, whose annotations are generated by the teacher. Experiments were conducted on five state-of-the-art networks for semantic segmentation and seven datasets across three imaging modalities. We studied knowledge transfer from a single teacher, combination of knowledge transfer and fine-tuning, and knowledge transfer from multiple teachers. The student model with a single teacher achieved similar performance as the teacher; and the student model with multiple teachers achieved better performance than the teachers. The salient features of our algorithm include: 1)no need for original training data or generative networks, 2) knowledge transfer between different architectures, 3) ease of implementation for downstream tasks by using the downstream task dataset as the transferal dataset, 4) knowledge transfer of an ensemble of models, trained independently, into one student model. Extensive experiments demonstrate that the proposed algorithm is effective for knowledge transfer and easily tunable.
U-Net Using Stacked Dilated Convolutions for Medical Image Segmentation
Apr 10, 2020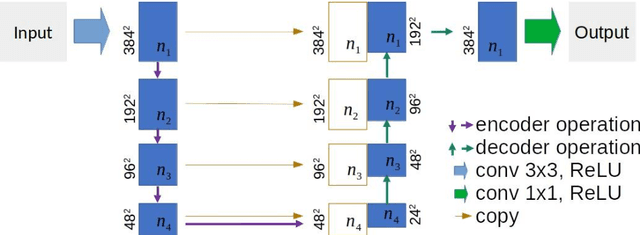

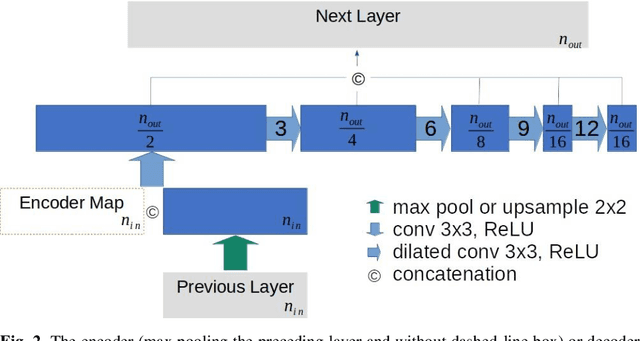
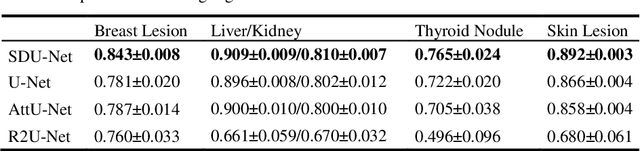
Abstract:This paper proposes a novel U-Net variant using stacked dilated convolutions for medical image segmentation (SDU-Net). SDU-Net adopts the architecture of vanilla U-Net with modifications in the encoder and decoder operations (an operation indicates all the processing for feature maps of the same resolution). Unlike vanilla U-Net which incorporates two standard convolutions in each encoder/decoder operation, SDU-Net uses one standard convolution followed by multiple dilated convolutions and concatenates all dilated convolution outputs as input to the next operation. Experiments showed that SDU-Net outperformed vanilla U-Net, attention U-Net (AttU-Net), and recurrent residual U-Net (R2U-Net) in all four tested segmentation tasks while using parameters around 40% of vanilla U-Net's, 17% of AttU-Net's, and 15% of R2U-Net's.
Weakly Supervised Context Encoder using DICOM metadata in Ultrasound Imaging
Mar 20, 2020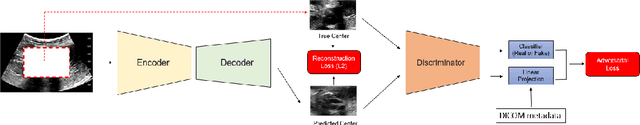
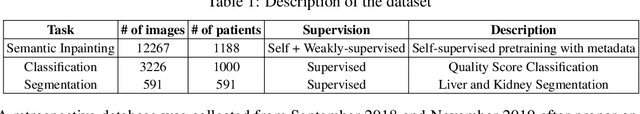
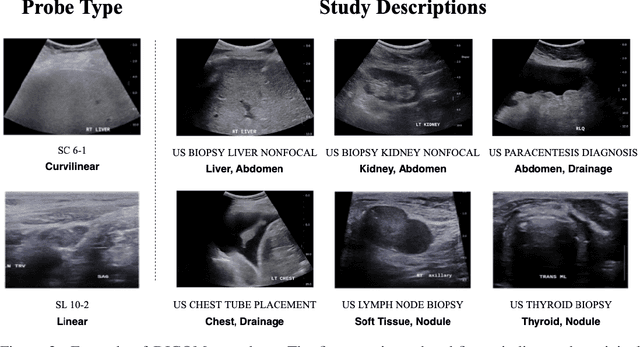
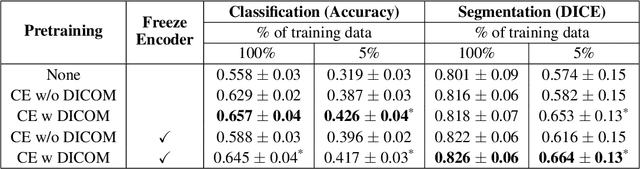
Abstract:Modern deep learning algorithms geared towards clinical adaption rely on a significant amount of high fidelity labeled data. Low-resource settings pose challenges like acquiring high fidelity data and becomes the bottleneck for developing artificial intelligence applications. Ultrasound images, stored in Digital Imaging and Communication in Medicine (DICOM) format, have additional metadata data corresponding to ultrasound image parameters and medical exams. In this work, we leverage DICOM metadata from ultrasound images to help learn representations of the ultrasound image. We demonstrate that the proposed method outperforms the non-metadata based approaches across different downstream tasks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge