Viksit Kumar
Efficient Aberration Correction via Optimal Bulk Speed of Sound Compensation
Mar 03, 2023



Abstract:Diagnostic ultrasound is a versatile and practical tool in the abdomen, and is particularly vital toward the detection and mitigation of early-stage non-alcoholic fatty liver disease (NAFLD). However, its performance in those with obesity -- who are at increased risk for NAFLD -- is degraded due to distortions of the ultrasound as it traverses thicker, acoustically heterogeneous body walls (aberration). Here, we assess the capability of a bulk speed of sound correction in receive beamforming to correct aberration, and improve the resulting images. We find that a bulk correction may approximate the aberration profile for layers or relevant thicknesses (1 to 3 cm) and speeds of sound (1400 to 1500 m/s). Additionally, through in vitro experiments, we show significant improvement in resolution (average point target width reduced by 60 %) with bulk speed of sound correction determined automatically from the beamformed images. Together, these results demonstrate the utility of simple, efficient bulk speed of sound correction to improve the quality of diagnostic liver images.
Network-Agnostic Knowledge Transfer for Medical Image Segmentation
Jan 23, 2021
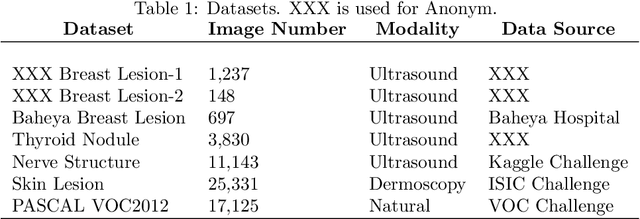
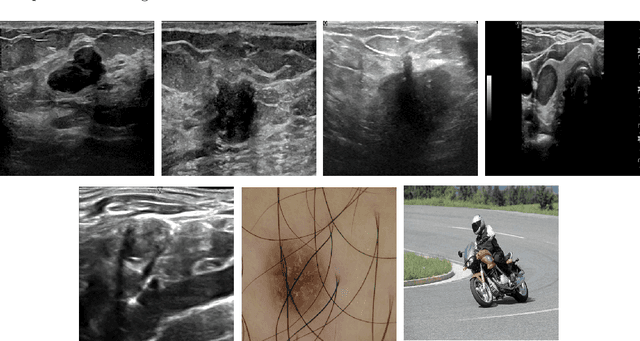
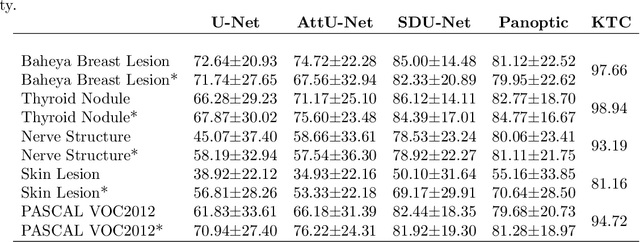
Abstract:Conventional transfer learning leverages weights of pre-trained networks, but mandates the need for similar neural architectures. Alternatively, knowledge distillation can transfer knowledge between heterogeneous networks but often requires access to the original training data or additional generative networks. Knowledge transfer between networks can be improved by being agnostic to the choice of network architecture and reducing the dependence on original training data. We propose a knowledge transfer approach from a teacher to a student network wherein we train the student on an independent transferal dataset, whose annotations are generated by the teacher. Experiments were conducted on five state-of-the-art networks for semantic segmentation and seven datasets across three imaging modalities. We studied knowledge transfer from a single teacher, combination of knowledge transfer and fine-tuning, and knowledge transfer from multiple teachers. The student model with a single teacher achieved similar performance as the teacher; and the student model with multiple teachers achieved better performance than the teachers. The salient features of our algorithm include: 1)no need for original training data or generative networks, 2) knowledge transfer between different architectures, 3) ease of implementation for downstream tasks by using the downstream task dataset as the transferal dataset, 4) knowledge transfer of an ensemble of models, trained independently, into one student model. Extensive experiments demonstrate that the proposed algorithm is effective for knowledge transfer and easily tunable.
U-Net Using Stacked Dilated Convolutions for Medical Image Segmentation
Apr 10, 2020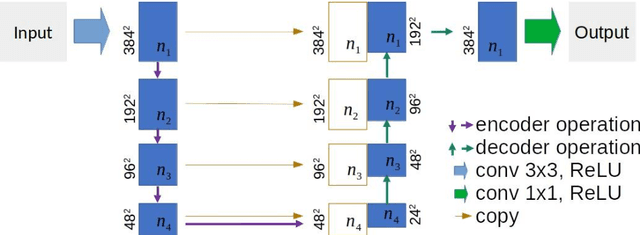

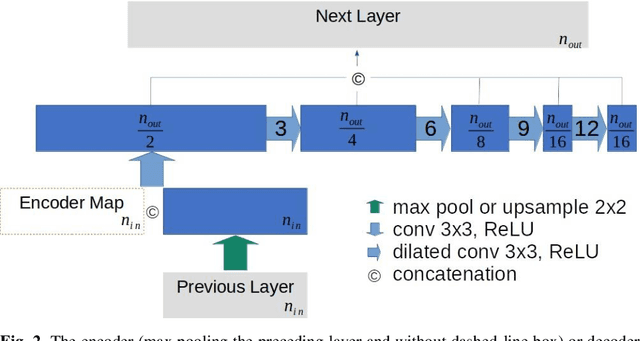
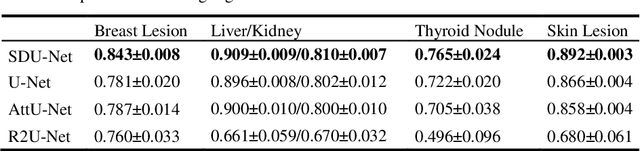
Abstract:This paper proposes a novel U-Net variant using stacked dilated convolutions for medical image segmentation (SDU-Net). SDU-Net adopts the architecture of vanilla U-Net with modifications in the encoder and decoder operations (an operation indicates all the processing for feature maps of the same resolution). Unlike vanilla U-Net which incorporates two standard convolutions in each encoder/decoder operation, SDU-Net uses one standard convolution followed by multiple dilated convolutions and concatenates all dilated convolution outputs as input to the next operation. Experiments showed that SDU-Net outperformed vanilla U-Net, attention U-Net (AttU-Net), and recurrent residual U-Net (R2U-Net) in all four tested segmentation tasks while using parameters around 40% of vanilla U-Net's, 17% of AttU-Net's, and 15% of R2U-Net's.
Weakly Supervised Context Encoder using DICOM metadata in Ultrasound Imaging
Mar 20, 2020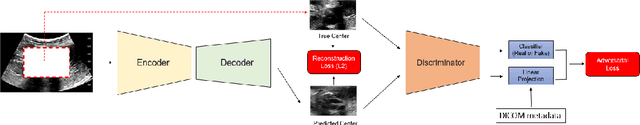
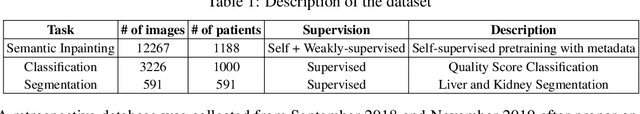
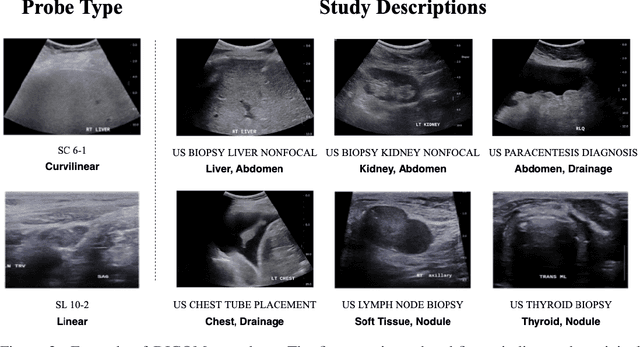
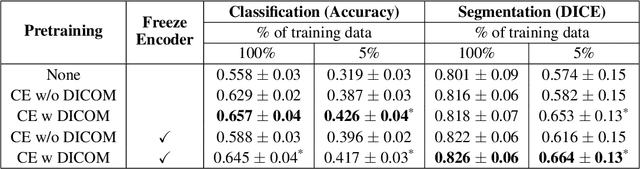
Abstract:Modern deep learning algorithms geared towards clinical adaption rely on a significant amount of high fidelity labeled data. Low-resource settings pose challenges like acquiring high fidelity data and becomes the bottleneck for developing artificial intelligence applications. Ultrasound images, stored in Digital Imaging and Communication in Medicine (DICOM) format, have additional metadata data corresponding to ultrasound image parameters and medical exams. In this work, we leverage DICOM metadata from ultrasound images to help learn representations of the ultrasound image. We demonstrate that the proposed method outperforms the non-metadata based approaches across different downstream tasks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge