Saurabh Deshpande
REVEAL: Multi-turn Evaluation of Image-Input Harms for Vision LLM
May 07, 2025Abstract:Vision Large Language Models (VLLMs) represent a significant advancement in artificial intelligence by integrating image-processing capabilities with textual understanding, thereby enhancing user interactions and expanding application domains. However, their increased complexity introduces novel safety and ethical challenges, particularly in multi-modal and multi-turn conversations. Traditional safety evaluation frameworks, designed for text-based, single-turn interactions, are inadequate for addressing these complexities. To bridge this gap, we introduce the REVEAL (Responsible Evaluation of Vision-Enabled AI LLMs) Framework, a scalable and automated pipeline for evaluating image-input harms in VLLMs. REVEAL includes automated image mining, synthetic adversarial data generation, multi-turn conversational expansion using crescendo attack strategies, and comprehensive harm assessment through evaluators like GPT-4o. We extensively evaluated five state-of-the-art VLLMs, GPT-4o, Llama-3.2, Qwen2-VL, Phi3.5V, and Pixtral, across three important harm categories: sexual harm, violence, and misinformation. Our findings reveal that multi-turn interactions result in significantly higher defect rates compared to single-turn evaluations, highlighting deeper vulnerabilities in VLLMs. Notably, GPT-4o demonstrated the most balanced performance as measured by our Safety-Usability Index (SUI) followed closely by Pixtral. Additionally, misinformation emerged as a critical area requiring enhanced contextual defenses. Llama-3.2 exhibited the highest MT defect rate ($16.55 \%$) while Qwen2-VL showed the highest MT refusal rate ($19.1 \%$).
Graph Representation Learning Strategies for Omics Data: A Case Study on Parkinson's Disease
Jun 20, 2024Abstract:Omics data analysis is crucial for studying complex diseases, but its high dimensionality and heterogeneity challenge classical statistical and machine learning methods. Graph neural networks have emerged as promising alternatives, yet the optimal strategies for their design and optimization in real-world biomedical challenges remain unclear. This study evaluates various graph representation learning models for case-control classification using high-throughput biological data from Parkinson's disease and control samples. We compare topologies derived from sample similarity networks and molecular interaction networks, including protein-protein and metabolite-metabolite interactions (PPI, MMI). Graph Convolutional Network (GCNs), Chebyshev spectral graph convolution (ChebyNet), and Graph Attention Network (GAT), are evaluated alongside advanced architectures like graph transformers, the graph U-net, and simpler models like multilayer perceptron (MLP). These models are systematically applied to transcriptomics and metabolomics data independently. Our comparative analysis highlights the benefits and limitations of various architectures in extracting patterns from omics data, paving the way for more accurate and interpretable models in biomedical research.
Convolution, aggregation and attention based deep neural networks for accelerating simulations in mechanics
Dec 01, 2022Abstract:Deep learning surrogate models are being increasingly used in accelerating scientific simulations as a replacement for costly conventional numerical techniques. However, their use remains a significant challenge when dealing with real-world complex examples. In this work, we demonstrate three types of neural network architectures for efficient learning of highly non-linear deformations of solid bodies. The first two architectures are based on the recently proposed CNN U-NET and MAgNET (graph U-NET) frameworks which have shown promising performance for learning on mesh-based data. The third architecture is Perceiver IO, a very recent architecture that belongs to the family of attention-based neural networks--a class that has revolutionised diverse engineering fields and is still unexplored in computational mechanics. We study and compare the performance of all three networks on two benchmark examples, and show their capabilities to accurately predict the non-linear mechanical responses of soft bodies.
MAgNET: A Graph U-Net Architecture for Mesh-Based Simulations
Nov 01, 2022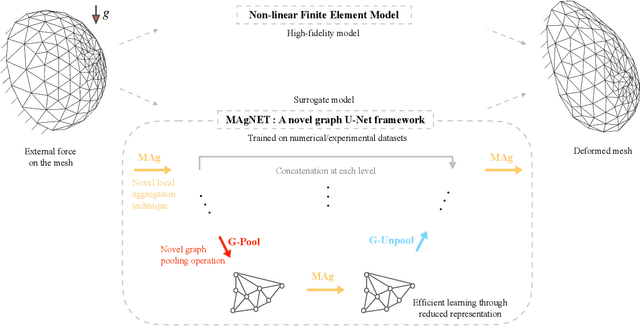
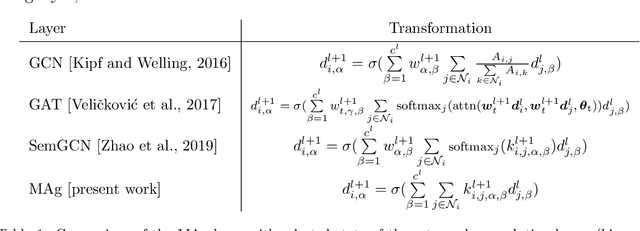
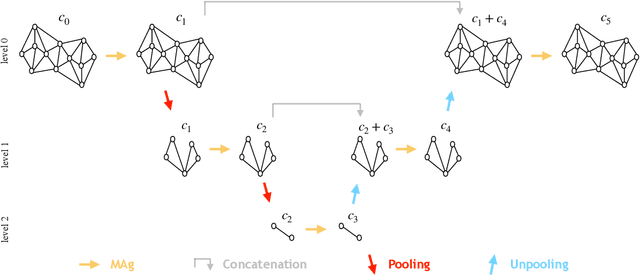
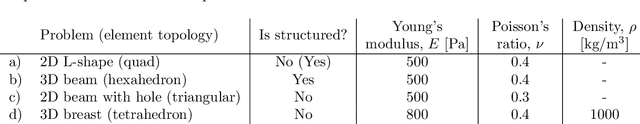
Abstract:Mesh-based approaches are fundamental to solving physics-based simulations, however, they require significant computational efforts, especially for highly non-linear problems. Deep learning techniques accelerate physics-based simulations, however, they fail to perform efficiently as the size and complexity of the problem increases. Hence in this work, we propose MAgNET: Multi-channel Aggregation Network, a novel geometric deep learning framework for performing supervised learning on mesh-based graph data. MAgNET is based on the proposed MAg (Multichannel Aggregation) operation which generalises the concept of multi-channel local operations in convolutional neural networks to arbitrary non-grid inputs. MAg can efficiently perform non-linear regression mapping for graph-structured data. MAg layers are interleaved with the proposed novel graph pooling operations to constitute a graph U-Net architecture that is robust, handles arbitrary complex meshes and scales efficiently with the size of the problem. Although not limited to the type of discretisation, we showcase the predictive capabilities of MAgNET for several non-linear finite element simulations.
FEM-based Real-Time Simulations of Large Deformations with Probabilistic Deep Learning
Nov 02, 2021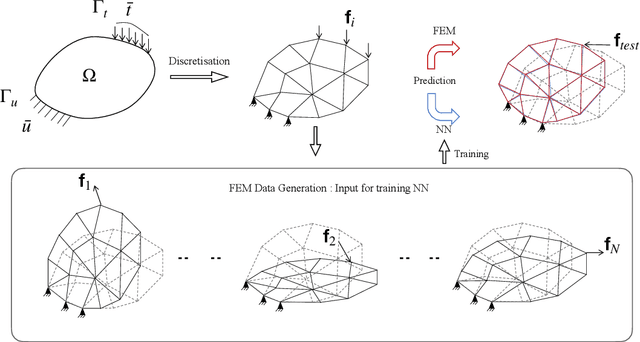
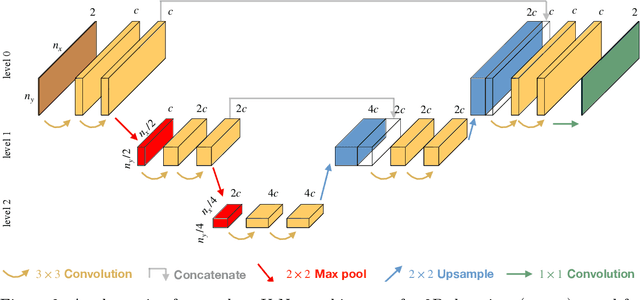

Abstract:For many engineering applications, such as real-time simulations or control, conventional solution techniques of the underlying nonlinear problems are usually computationally too expensive. In this work, we propose a highly efficient deep-learning surrogate framework that is able to predict the response of hyper-elastic bodies under load. The surrogate model takes the form of special convolutional neural network architecture, so-called U-Net, which is trained with force-displacement data obtained with the finite element method. We propose deterministic- and probabilistic versions of the framework and study it for three benchmark problems. In particular, we check the capabilities of the Maximum Likelihood and the Variational Bayes Inference formulations to assess the confidence intervals of solutions.
AutoKnow: Self-Driving Knowledge Collection for Products of Thousands of Types
Jun 24, 2020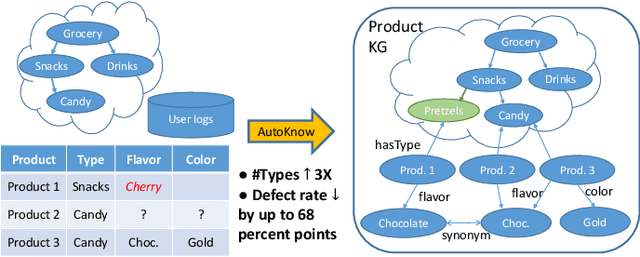
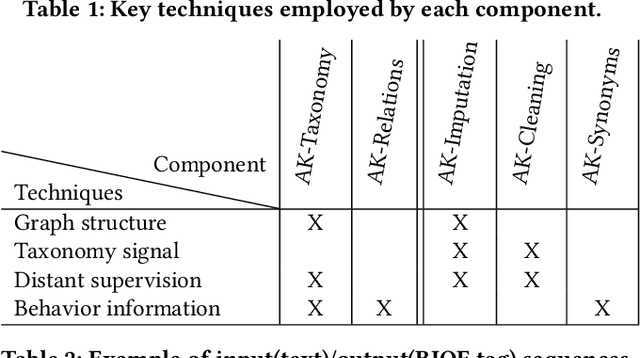
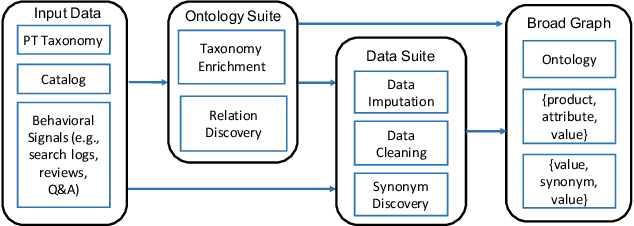
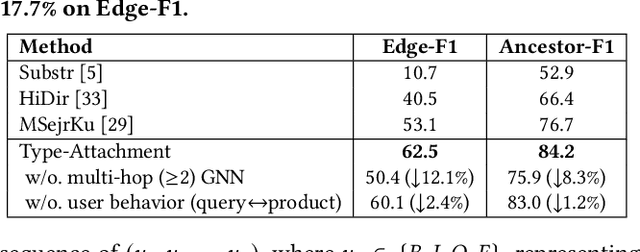
Abstract:Can one build a knowledge graph (KG) for all products in the world? Knowledge graphs have firmly established themselves as valuable sources of information for search and question answering, and it is natural to wonder if a KG can contain information about products offered at online retail sites. There have been several successful examples of generic KGs, but organizing information about products poses many additional challenges, including sparsity and noise of structured data for products, complexity of the domain with millions of product types and thousands of attributes, heterogeneity across large number of categories, as well as large and constantly growing number of products. We describe AutoKnow, our automatic (self-driving) system that addresses these challenges. The system includes a suite of novel techniques for taxonomy construction, product property identification, knowledge extraction, anomaly detection, and synonym discovery. AutoKnow is (a) automatic, requiring little human intervention, (b) multi-scalable, scalable in multiple dimensions (many domains, many products, and many attributes), and (c) integrative, exploiting rich customer behavior logs. AutoKnow has been operational in collecting product knowledge for over 11K product types.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge