Rosalind W. Picard
Improving Domain Generalization in Contrastive Learning using Adaptive Temperature Control
Jan 12, 2026Abstract:Self-supervised pre-training with contrastive learning is a powerful method for learning from sparsely labeled data. However, performance can drop considerably when there is a shift in the distribution of data from training to test time. We study this phenomenon in a setting in which the training data come from multiple domains, and the test data come from a domain not seen at training that is subject to significant covariate shift. We present a new method for contrastive learning that incorporates domain labels to increase the domain invariance of learned representations, leading to improved out-of-distribution generalization. Our method adjusts the temperature parameter in the InfoNCE loss -- which controls the relative weighting of negative pairs -- using the probability that a negative sample comes from the same domain as the anchor. This upweights pairs from more similar domains, encouraging the model to discriminate samples based on domain-invariant attributes. Through experiments on a variant of the MNIST dataset, we demonstrate that our method yields better out-of-distribution performance than domain generalization baselines. Furthermore, our method maintains strong in-distribution task performance, substantially outperforming baselines on this measure.
DISSECT: Disentangled Simultaneous Explanations via Concept Traversals
May 31, 2021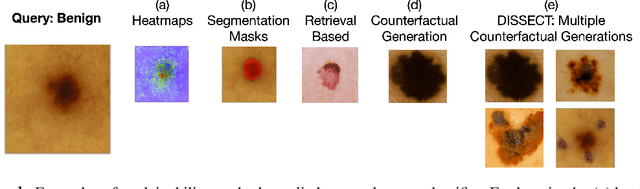
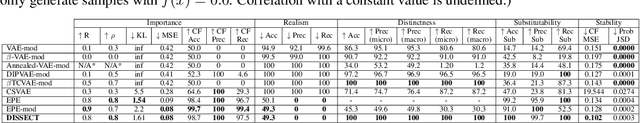
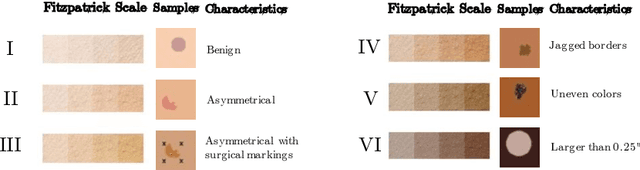
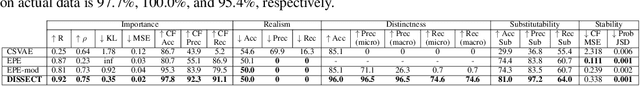
Abstract:Explaining deep learning model inferences is a promising venue for scientific understanding, improving safety, uncovering hidden biases, evaluating fairness, and beyond, as argued by many scholars. One of the principal benefits of counterfactual explanations is allowing users to explore "what-if" scenarios through what does not and cannot exist in the data, a quality that many other forms of explanation such as heatmaps and influence functions are inherently incapable of doing. However, most previous work on generative explainability cannot disentangle important concepts effectively, produces unrealistic examples, or fails to retain relevant information. We propose a novel approach, DISSECT, that jointly trains a generator, a discriminator, and a concept disentangler to overcome such challenges using little supervision. DISSECT generates Concept Traversals (CTs), defined as a sequence of generated examples with increasing degrees of concepts that influence a classifier's decision. By training a generative model from a classifier's signal, DISSECT offers a way to discover a classifier's inherent "notion" of distinct concepts automatically rather than rely on user-predefined concepts. We show that DISSECT produces CTs that (1) disentangle several concepts, (2) are influential to a classifier's decision and are coupled to its reasoning due to joint training (3), are realistic, (4) preserve relevant information, and (5) are stable across similar inputs. We validate DISSECT on several challenging synthetic and realistic datasets where previous methods fall short of satisfying desirable criteria for interpretability and show that it performs consistently well and better than existing methods. Finally, we present experiments showing applications of DISSECT for detecting potential biases of a classifier and identifying spurious artifacts that impact predictions.
Personalized Federated Deep Learning for Pain Estimation From Face Images
Jan 12, 2021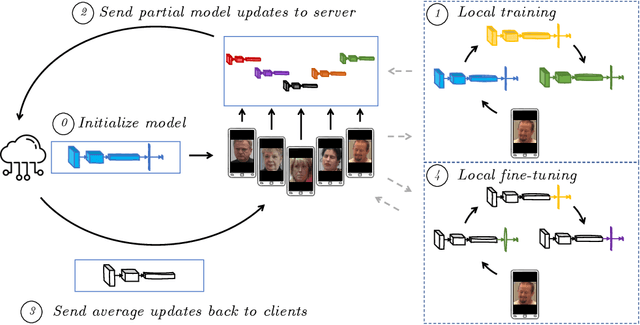
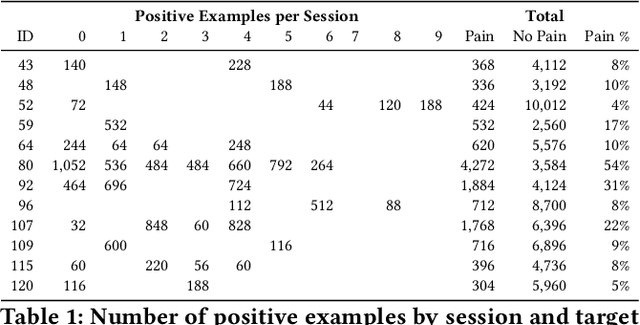
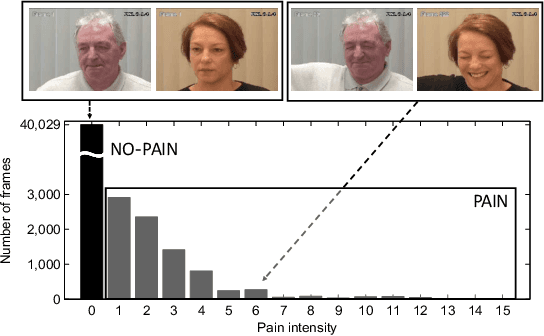

Abstract:Standard machine learning approaches require centralizing the users' data in one computer or a shared database, which raises data privacy and confidentiality concerns. Therefore, limiting central access is important, especially in healthcare settings, where data regulations are strict. A potential approach to tackling this is Federated Learning (FL), which enables multiple parties to collaboratively learn a shared prediction model by using parameters of locally trained models while keeping raw training data locally. In the context of AI-assisted pain-monitoring, we wish to enable confidentiality-preserving and unobtrusive pain estimation for long-term pain-monitoring and reduce the burden on the nursing staff who perform frequent routine check-ups. To this end, we propose a novel Personalized Federated Deep Learning (PFDL) approach for pain estimation from face images. PFDL performs collaborative training of a deep model, implemented using a lightweight CNN architecture, across different clients (i.e., subjects) without sharing their face images. Instead of sharing all parameters of the model, as in standard FL, PFDL retains the last layer locally (used to personalize the pain estimates). This (i) adds another layer of data confidentiality, making it difficult for an adversary to infer pain levels of the target subject, while (ii) personalizing the pain estimation to each subject through local parameter tuning. We show using a publicly available dataset of face videos of pain (UNBC-McMaster Shoulder Pain Database), that PFDL performs comparably or better than the standard centralized and FL algorithms, while further enhancing data privacy. This, has the potential to improve traditional pain monitoring by making it more secure, computationally efficient, and scalable to a large number of individuals (e.g., for in-home pain monitoring), providing timely and unobtrusive pain measurement.
openXDATA: A Tool for Multi-Target Data Generation and Missing Label Completion
Jul 27, 2020
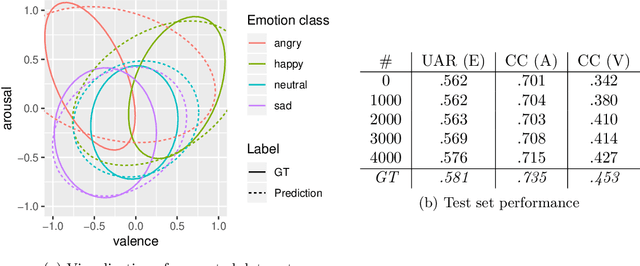
Abstract:A common problem in machine learning is to deal with datasets with disjoint label spaces and missing labels. In this work, we introduce the openXDATA tool that completes the missing labels in partially labelled or unlabelled datasets in order to generate multi-target data with labels in the joint label space of the datasets. To this end, we designed and implemented the cross-data label completion (CDLC) algorithm that uses a multi-task shared-hidden-layer DNN to iteratively complete the sparse label matrix of the instances from the different datasets. We apply the new tool to estimate labels across four emotion datasets: one labeled with discrete emotion categories (e.g., happy, sad, angry), one labeled with continuous values along arousal and valence dimensions, one with both kinds of labels, and one unlabeled. Testing with drop-out of true labels, we show the ability to estimate both categories and continuous labels for all of the datasets, at rates that approached the ground truth values. openXDATA is available under the GNU General Public License from https://github.com/fweninger/openXDATA.
Characterizing Sources of Uncertainty to Proxy Calibration and Disambiguate Annotator and Data Bias
Oct 05, 2019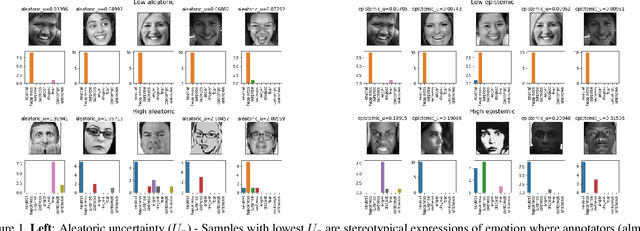
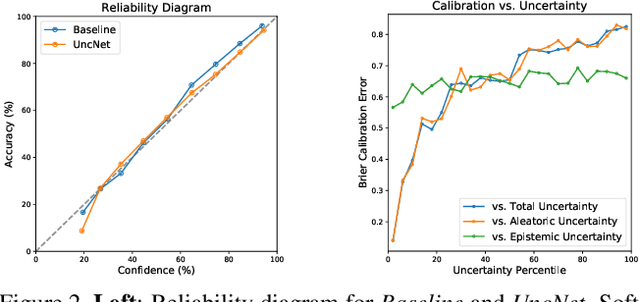
Abstract:Supporting model interpretability for complex phenomena where annotators can legitimately disagree, such as emotion recognition, is a challenging machine learning task. In this work, we show that explicitly quantifying the uncertainty in such settings has interpretability benefits. We use a simple modification of a classical network inference using Monte Carlo dropout to give measures of epistemic and aleatoric uncertainty. We identify a significant correlation between aleatoric uncertainty and human annotator disagreement ($r\approx.3$). Additionally, we demonstrate how difficult and subjective training samples can be identified using aleatoric uncertainty and how epistemic uncertainty can reveal data bias that could result in unfair predictions. We identify the total uncertainty as a suitable surrogate for model calibration, i.e. the degree we can trust model's predicted confidence. In addition to explainability benefits, we observe modest performance boosts from incorporating model uncertainty.
Multi-modal Active Learning From Human Data: A Deep Reinforcement Learning Approach
Jun 07, 2019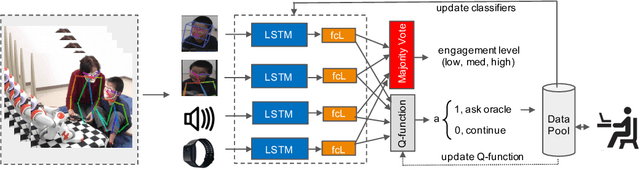
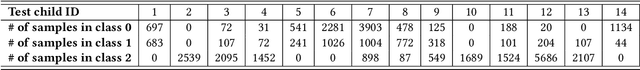
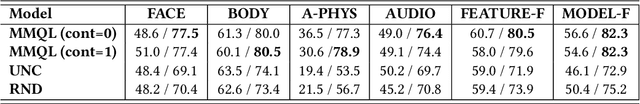
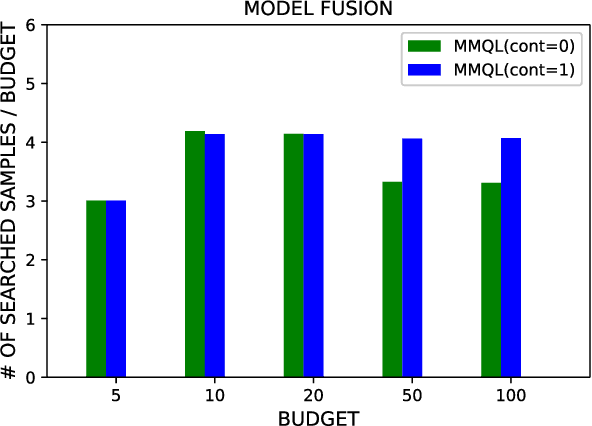
Abstract:Human behavior expression and experience are inherently multi-modal, and characterized by vast individual and contextual heterogeneity. To achieve meaningful human-computer and human-robot interactions, multi-modal models of the users states (e.g., engagement) are therefore needed. Most of the existing works that try to build classifiers for the users states assume that the data to train the models are fully labeled. Nevertheless, data labeling is costly and tedious, and also prone to subjective interpretations by the human coders. This is even more pronounced when the data are multi-modal (e.g., some users are more expressive with their facial expressions, some with their voice). Thus, building models that can accurately estimate the users states during an interaction is challenging. To tackle this, we propose a novel multi-modal active learning (AL) approach that uses the notion of deep reinforcement learning (RL) to find an optimal policy for active selection of the users data, needed to train the target (modality-specific) models. We investigate different strategies for multi-modal data fusion, and show that the proposed model-level fusion coupled with RL outperforms the feature-level and modality-specific models, and the naive AL strategies such as random sampling, and the standard heuristics such as uncertainty sampling. We show the benefits of this approach on the task of engagement estimation from real-world child-robot interactions during an autism therapy. Importantly, we show that the proposed multi-modal AL approach can be used to efficiently personalize the engagement classifiers to the target user using a small amount of actively selected users data.
Meta-Weighted Gaussian Process Experts for Personalized Forecasting of AD Cognitive Changes
Apr 19, 2019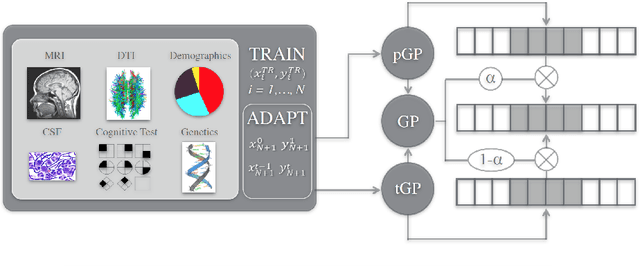
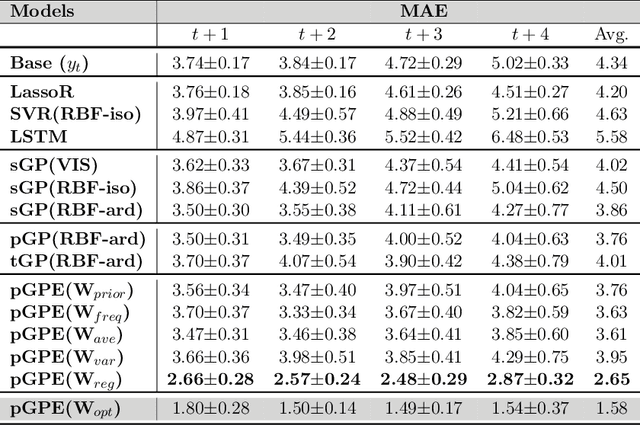
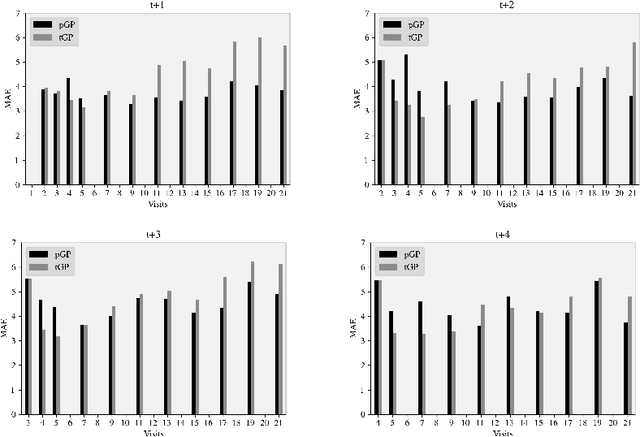
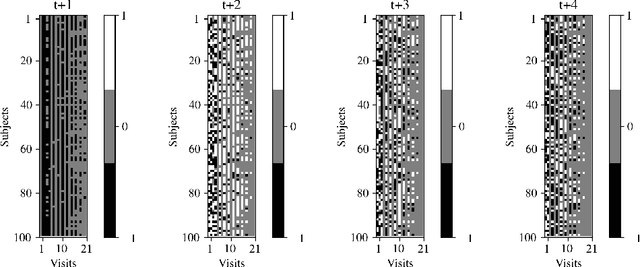
Abstract:We introduce a novel personalized Gaussian Process Experts (pGPE) model for predicting per-subject ADAS-Cog13 cognitive scores -- a significant predictor of Alzheimer's Disease (AD) in the cognitive domain -- over the future 6, 12, 18, and 24 months. We start by training a population-level model using multi-modal data from previously seen subjects using a base Gaussian Process (GP) regression. Then, we personalize this model by adapting the base GP sequentially over time to a new (target) subject using domain adaptive GPs, and also by training subject-specific GP. While we show that these models achieve improved performance when selectively applied to the forecasting task (one performs better than the other on different subjects/visits), the average performance per model is suboptimal. To this end, we used the notion of meta learning in the proposed pGPE to design a regression-based weighting of these expert models, where the expert weights are optimized for each subject and his/her future visit. The results on a cohort of subjects from the ADNI dataset show that this newly introduced personalized weighting of the expert models leads to large improvements in accurately forecasting future ADAS-Cog13 scores and their fine-grained changes associated with the AD progression. This approach has potential to help identify at-risk patients early and improve the construction of clinical trials for AD.
Estimating Carotid Pulse and Breathing Rate from Near-infrared Video of the Neck
May 24, 2018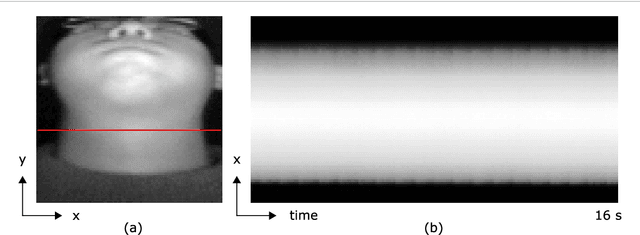
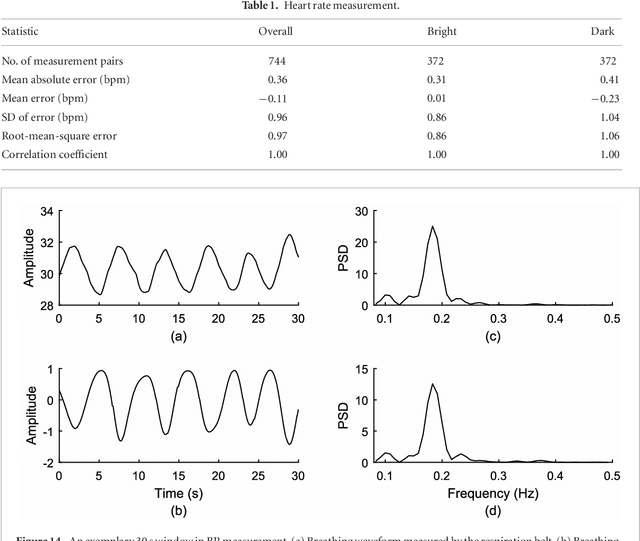
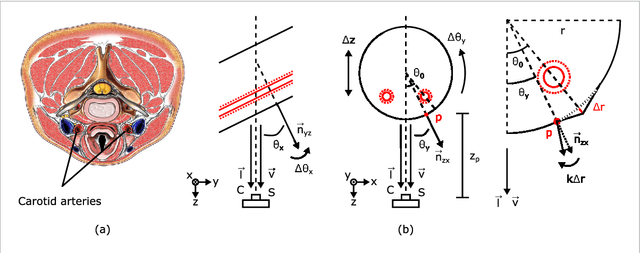
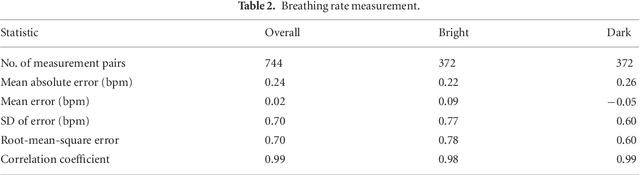
Abstract:Objective: Non-contact physiological measurement is a growing research area that allows capturing vital signs such as heart rate (HR) and breathing rate (BR) comfortably and unobtrusively with remote devices. However, most of the approaches work only in bright environments in which subtle photoplethysmographic and ballistocardiographic signals can be easily analyzed and/or require expensive and custom hardware to perform the measurements. Approach: This work introduces a low-cost method to measure subtle motions associated with the carotid pulse and breathing movement from the neck using near-infrared (NIR) video imaging. A skin reflection model of the neck was established to provide a theoretical foundation for the method. In particular, the method relies on template matching for neck detection, Principal Component Analysis for feature extraction, and Hidden Markov Models for data smoothing. Main Results: We compared the estimated HR and BR measures with ones provided by an FDA-cleared device in a 12-participant laboratory study: the estimates achieved a mean absolute error of 0.36 beats per minute and 0.24 breaths per minute under both bright and dark lighting. Significance: This work advances the possibilities of non-contact physiological measurement in real-life conditions in which environmental illumination is limited and in which the face of the person is not readily available or needs to be protected. Due to the increasing availability of NIR imaging devices, the described methods are readily scalable.
Personalized Gaussian Processes for Forecasting of Alzheimer's Disease Assessment Scale-Cognition Sub-Scale (ADAS-Cog13)
May 04, 2018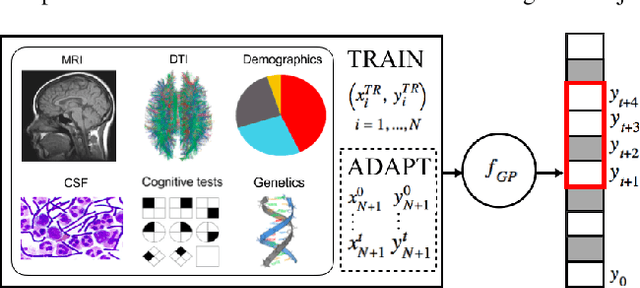
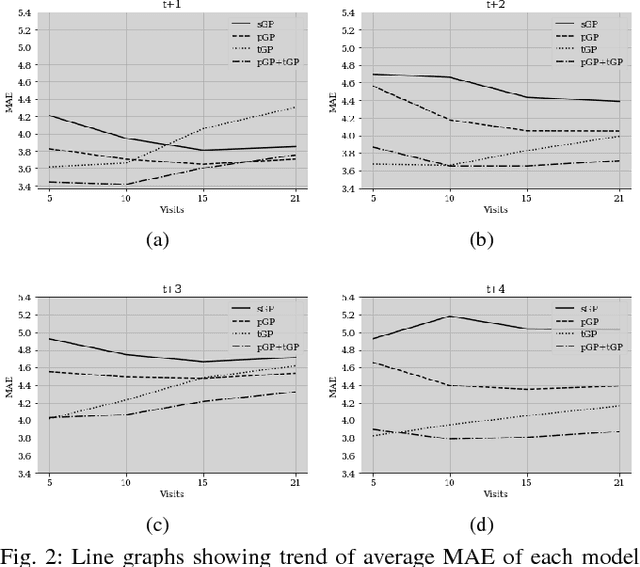

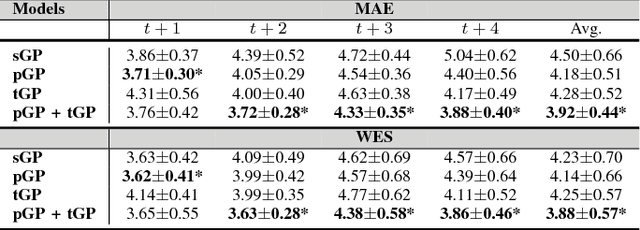
Abstract:In this paper, we introduce the use of a personalized Gaussian Process model (pGP) to predict per-patient changes in ADAS-Cog13 -- a significant predictor of Alzheimer's Disease (AD) in the cognitive domain -- using data from each patient's previous visits, and testing on future (held-out) data. We start by learning a population-level model using multi-modal data from previously seen patients using a base Gaussian Process (GP) regression. The personalized GP (pGP) is formed by adapting the base GP sequentially over time to a new (target) patient using domain adaptive GPs. We extend this personalized approach to predict the values of ADAS-Cog13 over the future 6, 12, 18, and 24 months. We compare this approach to a GP model trained only on past data of the target patients (tGP), as well as to a new approach that combines pGP with tGP. We find that the new approach, combining pGP with tGP, leads to large improvements in accurately forecasting future ADAS-Cog13 scores.
Personalized Gaussian Processes for Future Prediction of Alzheimer's Disease Progression
May 04, 2018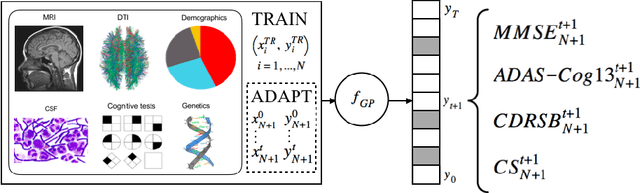
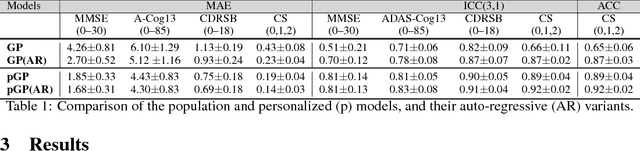

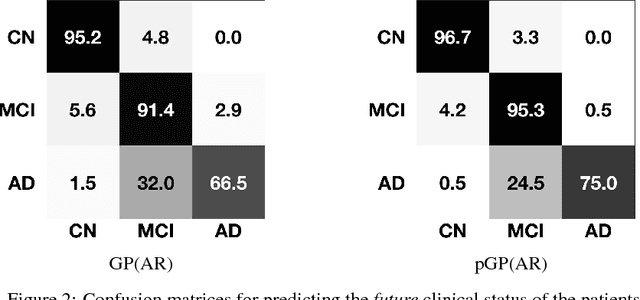
Abstract:In this paper, we introduce the use of a personalized Gaussian Process model (pGP) to predict the key metrics of Alzheimer's Disease progression (MMSE, ADAS-Cog13, CDRSB and CS) based on each patient's previous visits. We start by learning a population-level model using multi-modal data from previously seen patients using the base Gaussian Process (GP) regression. Then, this model is adapted sequentially over time to a new patient using domain adaptive GPs to form the patient's pGP. We show that this new approach, together with an auto-regressive formulation, leads to significant improvements in forecasting future clinical status and cognitive scores for target patients when compared to modeling the population with traditional GPs.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge