Romain Modzelewski
Predicting Patient Survival with Airway Biomarkers using nn-Unet/Radiomics
Jun 13, 2025Abstract:The primary objective of the AIIB 2023 competition is to evaluate the predictive significance of airway-related imaging biomarkers in determining the survival outcomes of patients with lung fibrosis.This study introduces a comprehensive three-stage approach. Initially, a segmentation network, namely nn-Unet, is employed to delineate the airway's structural boundaries. Subsequently, key features are extracted from the radiomic images centered around the trachea and an enclosing bounding box around the airway. This step is motivated by the potential presence of critical survival-related insights within the tracheal region as well as pertinent information encoded in the structure and dimensions of the airway. Lastly, radiomic features obtained from the segmented areas are integrated into an SVM classifier. We could obtain an overall-score of 0.8601 for the segmentation in Task 1 while 0.7346 for the classification in Task 2.
Fed-BioMed: Open, Transparent and Trusted Federated Learning for Real-world Healthcare Applications
Apr 24, 2023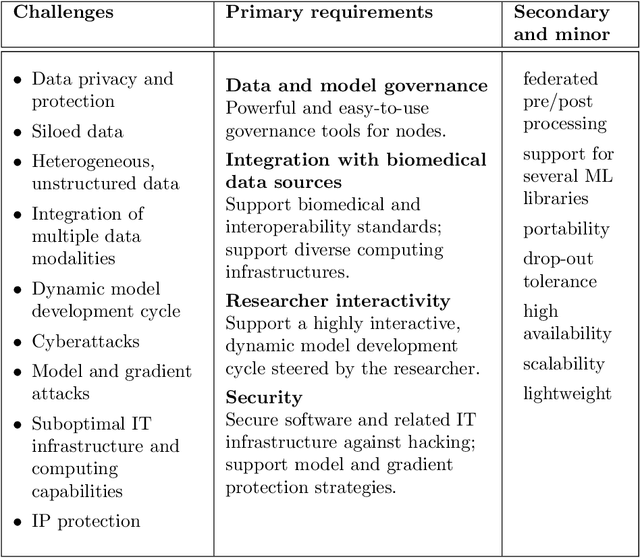
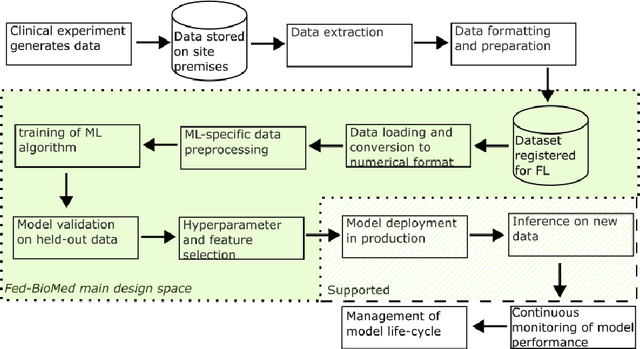
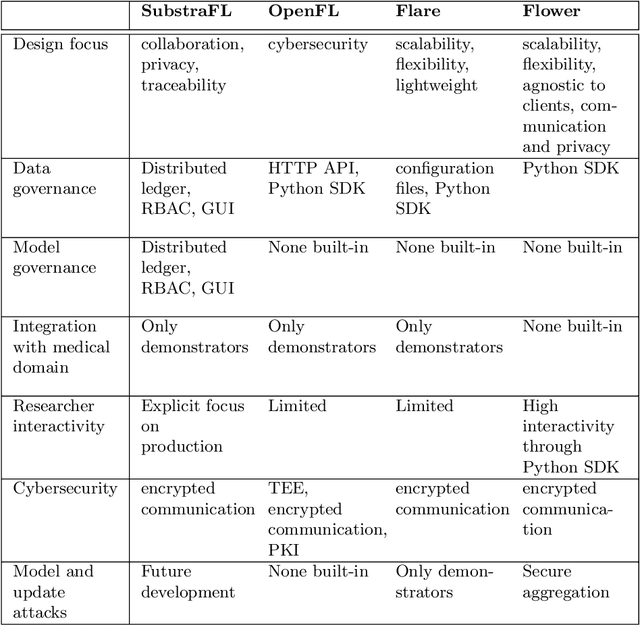
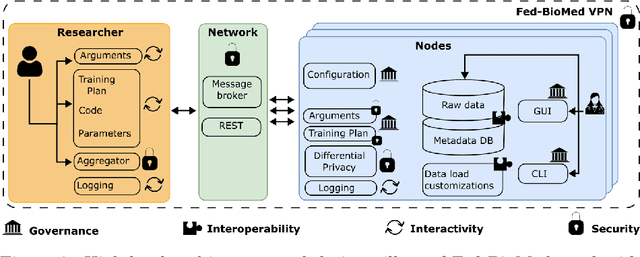
Abstract:The real-world implementation of federated learning is complex and requires research and development actions at the crossroad between different domains ranging from data science, to software programming, networking, and security. While today several FL libraries are proposed to data scientists and users, most of these frameworks are not designed to find seamless application in medical use-cases, due to the specific challenges and requirements of working with medical data and hospital infrastructures. Moreover, governance, design principles, and security assumptions of these frameworks are generally not clearly illustrated, thus preventing the adoption in sensitive applications. Motivated by the current technological landscape of FL in healthcare, in this document we present Fed-BioMed: a research and development initiative aiming at translating federated learning (FL) into real-world medical research applications. We describe our design space, targeted users, domain constraints, and how these factors affect our current and future software architecture.
Prediction of brain tumor recurrence location based on multi-modal fusion and nonlinear correlation learning
Apr 11, 2023Abstract:Brain tumor is one of the leading causes of cancer death. The high-grade brain tumors are easier to recurrent even after standard treatment. Therefore, developing a method to predict brain tumor recurrence location plays an important role in the treatment planning and it can potentially prolong patient's survival time. There is still little work to deal with this issue. In this paper, we present a deep learning-based brain tumor recurrence location prediction network. Since the dataset is usually small, we propose to use transfer learning to improve the prediction. We first train a multi-modal brain tumor segmentation network on the public dataset BraTS 2021. Then, the pre-trained encoder is transferred to our private dataset for extracting the rich semantic features. Following that, a multi-scale multi-channel feature fusion model and a nonlinear correlation learning module are developed to learn the effective features. The correlation between multi-channel features is modeled by a nonlinear equation. To measure the similarity between the distributions of original features of one modality and the estimated correlated features of another modality, we propose to use Kullback-Leibler divergence. Based on this divergence, a correlation loss function is designed to maximize the similarity between the two feature distributions. Finally, two decoders are constructed to jointly segment the present brain tumor and predict its future tumor recurrence location. To the best of our knowledge, this is the first work that can segment the present tumor and at the same time predict future tumor recurrence location, making the treatment planning more efficient and precise. The experimental results demonstrated the effectiveness of our proposed method to predict the brain tumor recurrence location from the limited dataset.
* 23 pages, 4 figures
Multi-Task Multi-Scale Learning For Outcome Prediction in 3D PET Images
Mar 01, 2022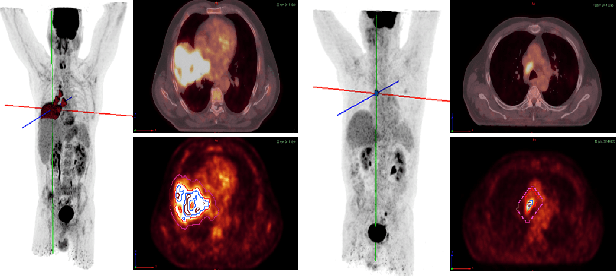
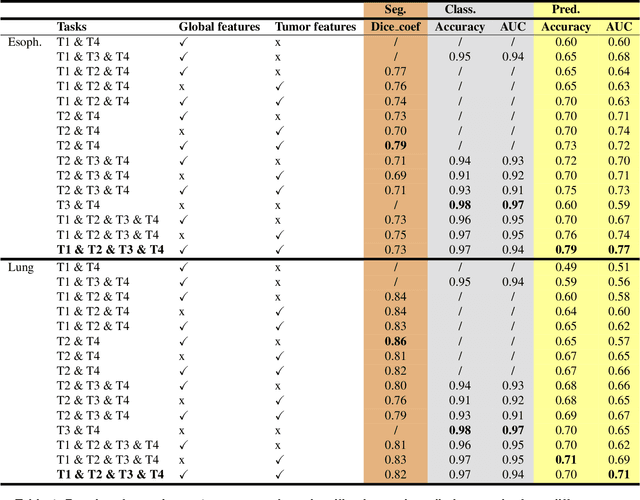
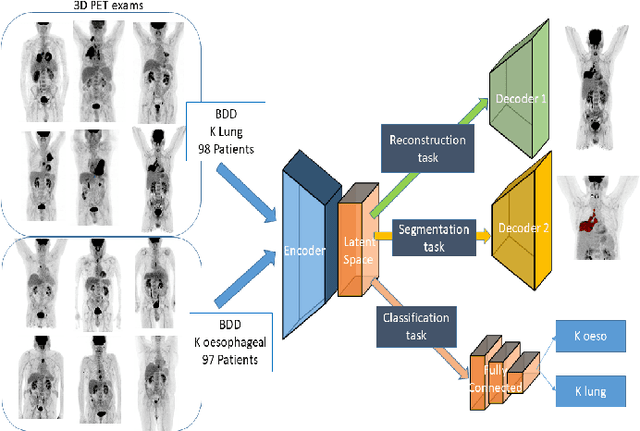
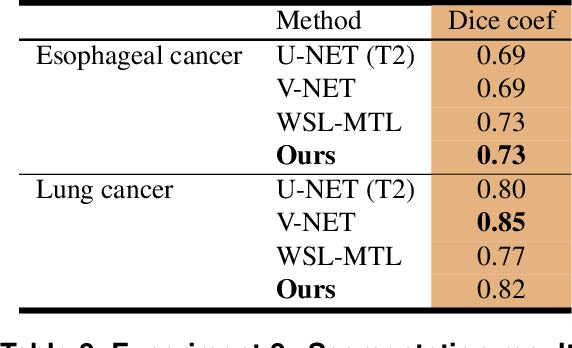
Abstract:Background and Objectives: Predicting patient response to treatment and survival in oncology is a prominent way towards precision medicine. To that end, radiomics was proposed as a field of study where images are used instead of invasive methods. The first step in radiomic analysis is the segmentation of the lesion. However, this task is time consuming and can be physician subjective. Automated tools based on supervised deep learning have made great progress to assist physicians. However, they are data hungry, and annotated data remains a major issue in the medical field where only a small subset of annotated images is available. Methods: In this work, we propose a multi-task learning framework to predict patient's survival and response. We show that the encoder can leverage multiple tasks to extract meaningful and powerful features that improve radiomics performance. We show also that subsidiary tasks serve as an inductive bias so that the model can better generalize. Results: Our model was tested and validated for treatment response and survival in lung and esophageal cancers, with an area under the ROC curve of 77% and 71% respectively, outperforming single task learning methods. Conclusions: We show that, by using a multi-task learning approach, we can boost the performance of radiomic analysis by extracting rich information of intratumoral and peritumoral regions.
RADIOGAN: Deep Convolutional Conditional Generative adversarial Network To Generate PET Images
Mar 19, 2020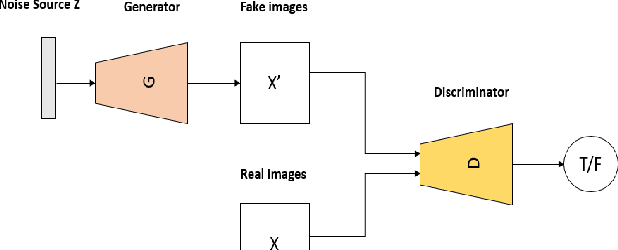
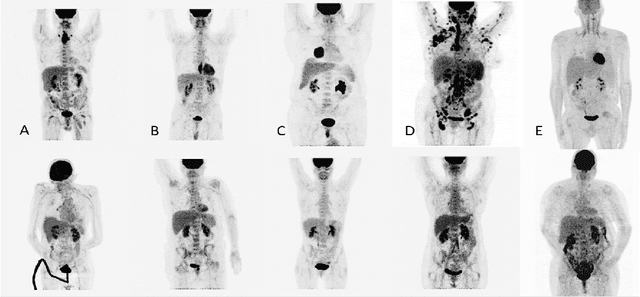
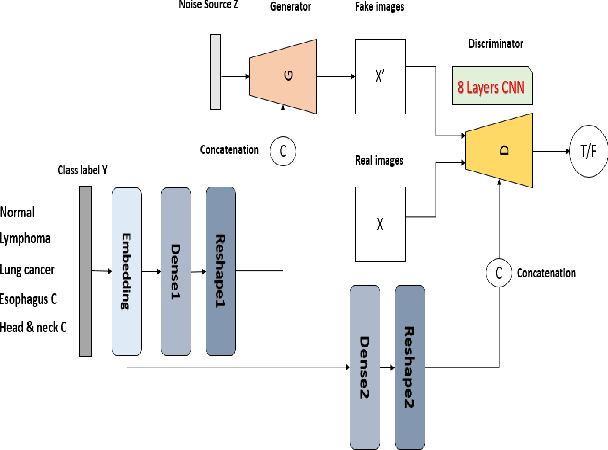
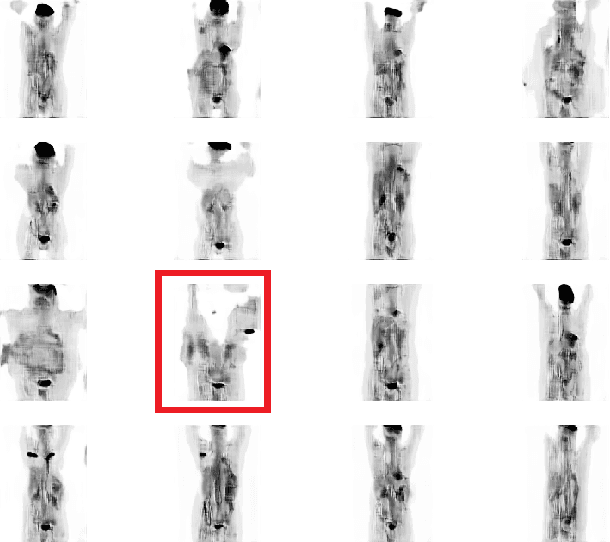
Abstract:One of the most challenges in medical imaging is the lack of data. It is proven that classical data augmentation methods are useful but still limited due to the huge variation in images. Using generative adversarial networks (GAN) is a promising way to address this problem, however, it is challenging to train one model to generate different classes of lesions. In this paper, we propose a deep convolutional conditional generative adversarial network to generate MIP positron emission tomography image (PET) which is a 2D image that represents a 3D volume for fast interpretation, according to different lesions or non lesion (normal). The advantage of our proposed method consists of one model that is capable of generating different classes of lesions trained on a small sample size for each class of lesion, and showing a very promising results. In addition, we show that a walk through a latent space can be used as a tool to evaluate the images generated.
Weakly Supervised PET Tumor Detection Using Class Response
Mar 19, 2020



Abstract:One of the most challenges in medical imaging is the lack of data and annotated data. It is proven that classical segmentation methods such as U-NET are useful but still limited due to the lack of annotated data. Using a weakly supervised learning is a promising way to address this problem, however, it is challenging to train one model to detect and locate efficiently different type of lesions due to the huge variation in images. In this paper, we present a novel approach to locate different type of lesions in positron emission tomography (PET) images using only a class label at the image-level. First, a simple convolutional neural network classifier is trained to predict the type of cancer on two 2D MIP images. Then, a pseudo-localization of the tumor is generated using class activation maps, back-propagated and corrected in a multitask learning approach with prior knowledge, resulting in a tumor detection mask. Finally, we use the mask generated from the two 2D images to detect the tumor in the 3D image. The advantage of our proposed method consists of detecting the whole tumor volume in 3D images, using only two 2D images of PET image, and showing a very promising results. It can be used as a tool to locate very efficiently tumors in a PET scan, which is a time-consuming task for physicians. In addition, we show that our proposed method can be used to conduct a radiomics study with state of the art results.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge