Tsiry Mayet
Predicting Patient Survival with Airway Biomarkers using nn-Unet/Radiomics
Jun 13, 2025Abstract:The primary objective of the AIIB 2023 competition is to evaluate the predictive significance of airway-related imaging biomarkers in determining the survival outcomes of patients with lung fibrosis.This study introduces a comprehensive three-stage approach. Initially, a segmentation network, namely nn-Unet, is employed to delineate the airway's structural boundaries. Subsequently, key features are extracted from the radiomic images centered around the trachea and an enclosing bounding box around the airway. This step is motivated by the potential presence of critical survival-related insights within the tracheal region as well as pertinent information encoded in the structure and dimensions of the airway. Lastly, radiomic features obtained from the segmented areas are integrated into an SVM classifier. We could obtain an overall-score of 0.8601 for the segmentation in Task 1 while 0.7346 for the classification in Task 2.
TD-Paint: Faster Diffusion Inpainting Through Time Aware Pixel Conditioning
Oct 11, 2024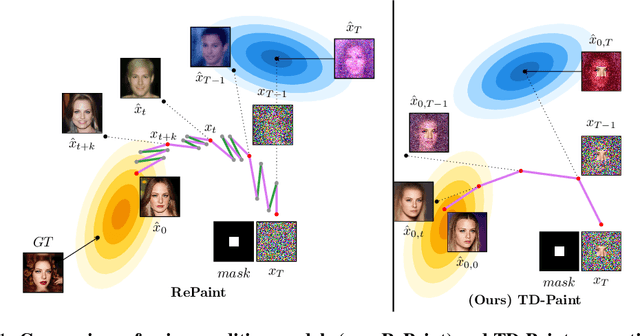

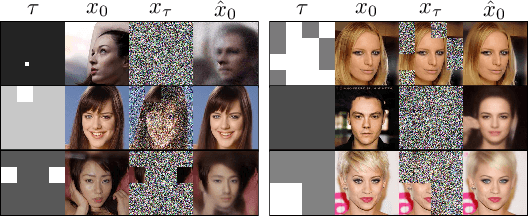

Abstract:Diffusion models have emerged as highly effective techniques for inpainting, however, they remain constrained by slow sampling rates. While recent advances have enhanced generation quality, they have also increased sampling time, thereby limiting scalability in real-world applications. We investigate the generative sampling process of diffusion-based inpainting models and observe that these models make minimal use of the input condition during the initial sampling steps. As a result, the sampling trajectory deviates from the data manifold, requiring complex synchronization mechanisms to realign the generation process. To address this, we propose Time-aware Diffusion Paint (TD-Paint), a novel approach that adapts the diffusion process by modeling variable noise levels at the pixel level. This technique allows the model to efficiently use known pixel values from the start, guiding the generation process toward the target manifold. By embedding this information early in the diffusion process, TD-Paint significantly accelerates sampling without compromising image quality. Unlike conventional diffusion-based inpainting models, which require a dedicated architecture or an expensive generation loop, TD-Paint achieves faster sampling times without architectural modifications. Experimental results across three datasets show that TD-Paint outperforms state-of-the-art diffusion models while maintaining lower complexity.
Source-Free Domain Adaptation of Weakly-Supervised Object Localization Models for Histology
Apr 29, 2024Abstract:Given the emergence of deep learning, digital pathology has gained popularity for cancer diagnosis based on histology images. Deep weakly supervised object localization (WSOL) models can be trained to classify histology images according to cancer grade and identify regions of interest (ROIs) for interpretation, using inexpensive global image-class annotations. A WSOL model initially trained on some labeled source image data can be adapted using unlabeled target data in cases of significant domain shifts caused by variations in staining, scanners, and cancer type. In this paper, we focus on source-free (unsupervised) domain adaptation (SFDA), a challenging problem where a pre-trained source model is adapted to a new target domain without using any source domain data for privacy and efficiency reasons. SFDA of WSOL models raises several challenges in histology, most notably because they are not intended to adapt for both classification and localization tasks. In this paper, 4 state-of-the-art SFDA methods, each one representative of a main SFDA family, are compared for WSOL in terms of classification and localization accuracy. They are the SFDA-Distribution Estimation, Source HypOthesis Transfer, Cross-Domain Contrastive Learning, and Adaptively Domain Statistics Alignment. Experimental results on the challenging Glas (smaller, breast cancer) and Camelyon16 (larger, colon cancer) histology datasets indicate that these SFDA methods typically perform poorly for localization after adaptation when optimized for classification.
Hunting imaging biomarkers in pulmonary fibrosis: Benchmarks of the AIIB23 challenge
Dec 21, 2023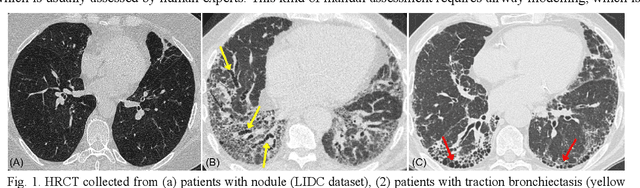
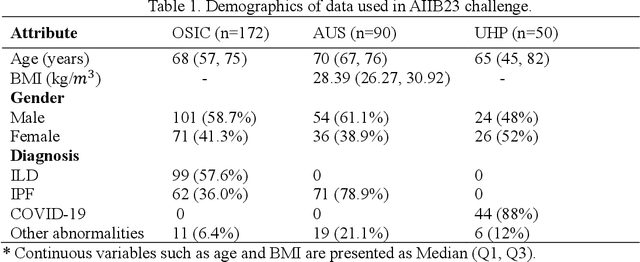
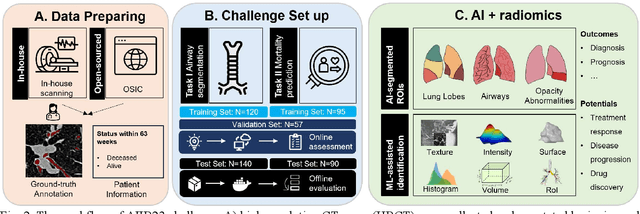
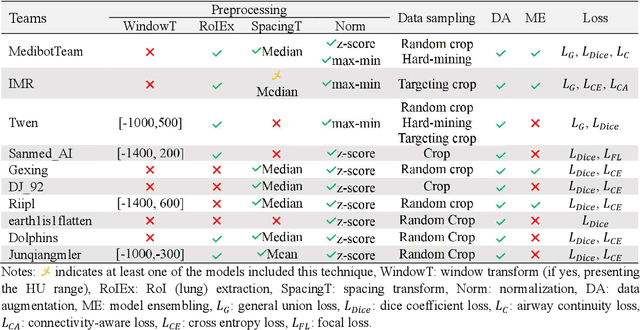
Abstract:Airway-related quantitative imaging biomarkers are crucial for examination, diagnosis, and prognosis in pulmonary diseases. However, the manual delineation of airway trees remains prohibitively time-consuming. While significant efforts have been made towards enhancing airway modelling, current public-available datasets concentrate on lung diseases with moderate morphological variations. The intricate honeycombing patterns present in the lung tissues of fibrotic lung disease patients exacerbate the challenges, often leading to various prediction errors. To address this issue, the 'Airway-Informed Quantitative CT Imaging Biomarker for Fibrotic Lung Disease 2023' (AIIB23) competition was organized in conjunction with the official 2023 International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI). The airway structures were meticulously annotated by three experienced radiologists. Competitors were encouraged to develop automatic airway segmentation models with high robustness and generalization abilities, followed by exploring the most correlated QIB of mortality prediction. A training set of 120 high-resolution computerised tomography (HRCT) scans were publicly released with expert annotations and mortality status. The online validation set incorporated 52 HRCT scans from patients with fibrotic lung disease and the offline test set included 140 cases from fibrosis and COVID-19 patients. The results have shown that the capacity of extracting airway trees from patients with fibrotic lung disease could be enhanced by introducing voxel-wise weighted general union loss and continuity loss. In addition to the competitive image biomarkers for prognosis, a strong airway-derived biomarker (Hazard ratio>1.5, p<0.0001) was revealed for survival prognostication compared with existing clinical measurements, clinician assessment and AI-based biomarkers.
Multiple Noises in Diffusion Model for Semi-Supervised Multi-Domain Translation
Sep 25, 2023Abstract:Domain-to-domain translation involves generating a target domain sample given a condition in the source domain. Most existing methods focus on fixed input and output domains, i.e. they only work for specific configurations (i.e. for two domains, either $D_1\rightarrow{}D_2$ or $D_2\rightarrow{}D_1$). This paper proposes Multi-Domain Diffusion (MDD), a conditional diffusion framework for multi-domain translation in a semi-supervised context. Unlike previous methods, MDD does not require defining input and output domains, allowing translation between any partition of domains within a set (such as $(D_1, D_2)\rightarrow{}D_3$, $D_2\rightarrow{}(D_1, D_3)$, $D_3\rightarrow{}D_1$, etc. for 3 domains), without the need to train separate models for each domain configuration. The key idea behind MDD is to leverage the noise formulation of diffusion models by incorporating one noise level per domain, which allows missing domains to be modeled with noise in a natural way. This transforms the training task from a simple reconstruction task to a domain translation task, where the model relies on less noisy domains to reconstruct more noisy domains. We present results on a multi-domain (with more than two domains) synthetic image translation dataset with challenging semantic domain inversion.
Domain Translation via Latent Space Mapping
Dec 06, 2022Abstract:In this paper, we investigate the problem of multi-domain translation: given an element $a$ of domain $A$, we would like to generate a corresponding $b$ sample in another domain $B$, and vice versa. Acquiring supervision in multiple domains can be a tedious task, also we propose to learn this translation from one domain to another when supervision is available as a pair $(a,b)\sim A\times B$ and leveraging possible unpaired data when only $a\sim A$ or only $b\sim B$ is available. We introduce a new unified framework called Latent Space Mapping (\model) that exploits the manifold assumption in order to learn, from each domain, a latent space. Unlike existing approaches, we propose to further regularize each latent space using available domains by learning each dependency between pairs of domains. We evaluate our approach in three tasks performing i) synthetic dataset with image translation, ii) real-world task of semantic segmentation for medical images, and iii) real-world task of facial landmark detection.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge