Luke McCaffrey
Goodman Cancer Research Centre Dept. of Oncology McGill University Canada
CLIP-IT: CLIP-based Pairing for Histology Images Classification
Apr 22, 2025Abstract:Multimodal learning has shown significant promise for improving medical image analysis by integrating information from complementary data sources. This is widely employed for training vision-language models (VLMs) for cancer detection based on histology images and text reports. However, one of the main limitations in training these VLMs is the requirement for large paired datasets, raising concerns over privacy, and data collection, annotation, and maintenance costs. To address this challenge, we introduce CLIP-IT method to train a vision backbone model to classify histology images by pairing them with privileged textual information from an external source. At first, the modality pairing step relies on a CLIP-based model to match histology images with semantically relevant textual report data from external sources, creating an augmented multimodal dataset without the need for manually paired samples. Then, we propose a multimodal training procedure that distills the knowledge from the paired text modality to the unimodal image classifier for enhanced performance without the need for the textual data during inference. A parameter-efficient fine-tuning method is used to efficiently address the misalignment between the main (image) and paired (text) modalities. During inference, the improved unimodal histology classifier is used, with only minimal additional computational complexity. Our experiments on challenging PCAM, CRC, and BACH histology image datasets show that CLIP-IT can provide a cost-effective approach to leverage privileged textual information and outperform unimodal classifiers for histology.
PixelCAM: Pixel Class Activation Mapping for Histology Image Classification and ROI Localization
Mar 31, 2025Abstract:Weakly supervised object localization (WSOL) methods allow training models to classify images and localize ROIs. WSOL only requires low-cost image-class annotations yet provides a visually interpretable classifier, which is important in histology image analysis. Standard WSOL methods rely on class activation mapping (CAM) methods to produce spatial localization maps according to a single- or two-step strategy. While both strategies have made significant progress, they still face several limitations with histology images. Single-step methods can easily result in under- or over-activation due to the limited visual ROI saliency in histology images and the limited localization cues. They also face the well-known issue of asynchronous convergence between classification and localization tasks. The two-step approach is sub-optimal because it is tied to a frozen classifier, limiting the capacity for localization. Moreover, these methods also struggle when applied to out-of-distribution (OOD) datasets. In this paper, a multi-task approach for WSOL is introduced for simultaneous training of both tasks to address the asynchronous convergence problem. In particular, localization is performed in the pixel-feature space of an image encoder that is shared with classification. This allows learning discriminant features and accurate delineation of foreground/background regions to support ROI localization and image classification. We propose PixelCAM, a cost-effective foreground/background pixel-wise classifier in the pixel-feature space that allows for spatial object localization. PixelCAM is trained using pixel pseudo-labels collected from a pretrained WSOL model. Both image and pixel-wise classifiers are trained simultaneously using standard gradient descent. In addition, our pixel classifier can easily be integrated into CNN- and transformer-based architectures without any modifications.
SR-CACO-2: A Dataset for Confocal Fluorescence Microscopy Image Super-Resolution
Jun 13, 2024



Abstract:Confocal fluorescence microscopy is one of the most accessible and widely used imaging techniques for the study of biological processes. Scanning confocal microscopy allows the capture of high-quality images from 3D samples, yet suffers from well-known limitations such as photobleaching and phototoxicity of specimens caused by intense light exposure, which limits its use in some applications, especially for living cells. Cellular damage can be alleviated by changing imaging parameters to reduce light exposure, often at the expense of image quality. Machine/deep learning methods for single-image super-resolution (SISR) can be applied to restore image quality by upscaling lower-resolution (LR) images to produce high-resolution images (HR). These SISR methods have been successfully applied to photo-realistic images due partly to the abundance of publicly available data. In contrast, the lack of publicly available data partly limits their application and success in scanning confocal microscopy. In this paper, we introduce a large scanning confocal microscopy dataset named SR-CACO-2 that is comprised of low- and high-resolution image pairs marked for three different fluorescent markers. It allows the evaluation of performance of SISR methods on three different upscaling levels (X2, X4, X8). SR-CACO-2 contains the human epithelial cell line Caco-2 (ATCC HTB-37), and it is composed of 22 tiles that have been translated in the form of 9,937 image patches for experiments with SISR methods. Given the new SR-CACO-2 dataset, we also provide benchmarking results for 15 state-of-the-art methods that are representative of the main SISR families. Results show that these methods have limited success in producing high-resolution textures, indicating that SR-CACO-2 represents a challenging problem. Our dataset, code and pretrained weights are available: https://github.com/sbelharbi/sr-caco-2.
Source-Free Domain Adaptation of Weakly-Supervised Object Localization Models for Histology
Apr 29, 2024Abstract:Given the emergence of deep learning, digital pathology has gained popularity for cancer diagnosis based on histology images. Deep weakly supervised object localization (WSOL) models can be trained to classify histology images according to cancer grade and identify regions of interest (ROIs) for interpretation, using inexpensive global image-class annotations. A WSOL model initially trained on some labeled source image data can be adapted using unlabeled target data in cases of significant domain shifts caused by variations in staining, scanners, and cancer type. In this paper, we focus on source-free (unsupervised) domain adaptation (SFDA), a challenging problem where a pre-trained source model is adapted to a new target domain without using any source domain data for privacy and efficiency reasons. SFDA of WSOL models raises several challenges in histology, most notably because they are not intended to adapt for both classification and localization tasks. In this paper, 4 state-of-the-art SFDA methods, each one representative of a main SFDA family, are compared for WSOL in terms of classification and localization accuracy. They are the SFDA-Distribution Estimation, Source HypOthesis Transfer, Cross-Domain Contrastive Learning, and Adaptively Domain Statistics Alignment. Experimental results on the challenging Glas (smaller, breast cancer) and Camelyon16 (larger, colon cancer) histology datasets indicate that these SFDA methods typically perform poorly for localization after adaptation when optimized for classification.
CoLo-CAM: Class Activation Mapping for Object Co-Localization in Weakly-Labeled Unconstrained Videos
Mar 16, 2023Abstract:Weakly-supervised video object localization (WSVOL) methods often rely on visual and motion cues only, making them susceptible to inaccurate localization. Recently, discriminative models via a temporal class activation mapping (CAM) method have been explored. Although results are promising, objects are assumed to have minimal movement leading to degradation in performance for relatively long-term dependencies. In this paper, a novel CoLo-CAM method for object localization is proposed to leverage spatiotemporal information in activation maps without any assumptions about object movement. Over a given sequence of frames, explicit joint learning of localization is produced across these maps based on color cues, by assuming an object has similar color across frames. The CAMs' activations are constrained to activate similarly over pixels with similar colors, achieving co-localization. This joint learning creates direct communication among pixels across all image locations, and over all frames, allowing for transfer, aggregation, and correction of learned localization. This is achieved by minimizing a color term of a CRF loss over joint images/maps. In addition to our multi-frame constraint, we impose per-frame local constraints including pseudo-labels, and CRF loss in combination with a global size constraint to improve per-frame localization. Empirical experiments on two challenging datasets for unconstrained videos, YouTube-Objects, show the merits of our method, and its robustness to long-term dependencies, leading to new state-of-the-art localization performance. Public code: https://github.com/sbelharbi/colo-cam.
TCAM: Temporal Class Activation Maps for Object Localization in Weakly-Labeled Unconstrained Videos
Aug 30, 2022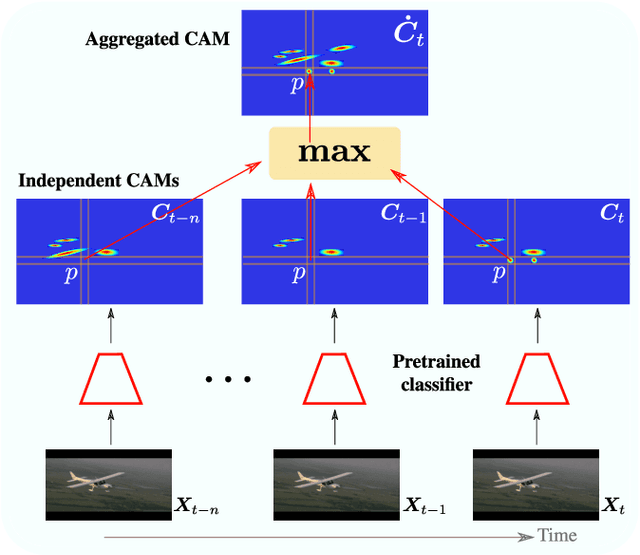
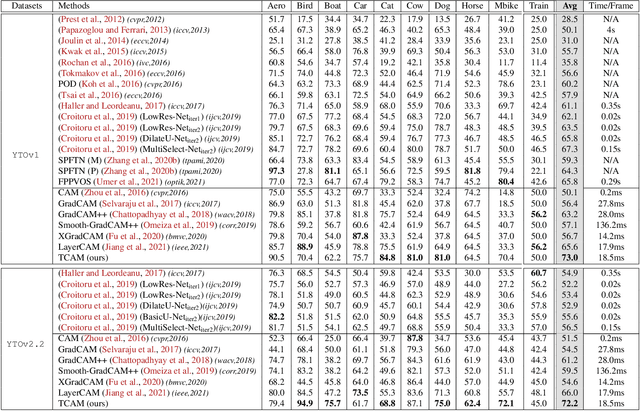
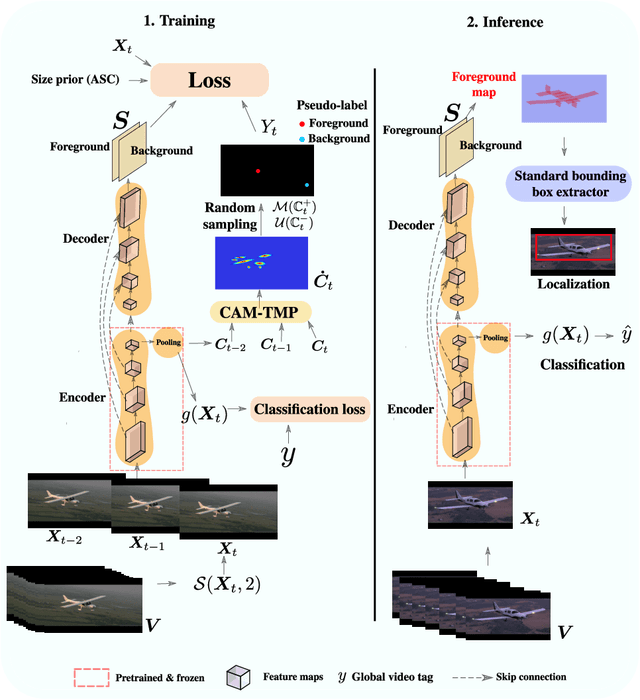
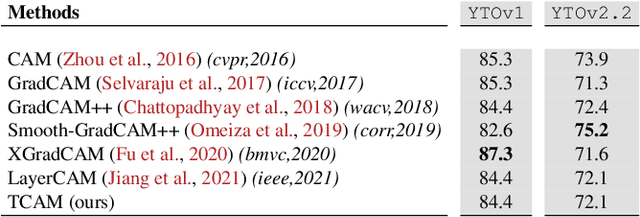
Abstract:Weakly supervised video object localization (WSVOL) allows locating object in videos using only global video tags such as object class. State-of-art methods rely on multiple independent stages, where initial spatio-temporal proposals are generated using visual and motion cues, then prominent objects are identified and refined. Localization is done by solving an optimization problem over one or more videos, and video tags are typically used for video clustering. This requires a model per-video or per-class making for costly inference. Moreover, localized regions are not necessary discriminant because of unsupervised motion methods like optical flow, or because video tags are discarded from optimization. In this paper, we leverage the successful class activation mapping (CAM) methods, designed for WSOL based on still images. A new Temporal CAM (TCAM) method is introduced to train a discriminant deep learning (DL) model to exploit spatio-temporal information in videos, using an aggregation mechanism, called CAM-Temporal Max Pooling (CAM-TMP), over consecutive CAMs. In particular, activations of regions of interest (ROIs) are collected from CAMs produced by a pretrained CNN classifier to build pixel-wise pseudo-labels for training the DL model. In addition, a global unsupervised size constraint, and local constraint such as CRF are used to yield more accurate CAMs. Inference over single independent frames allows parallel processing of a clip of frames, and real-time localization. Extensive experiments on two challenging YouTube-Objects datasets for unconstrained videos, indicate that CAM methods (trained on independent frames) can yield decent localization accuracy. Our proposed TCAM method achieves a new state-of-art in WSVOL accuracy, and visual results suggest that it can be adapted for subsequent tasks like visual object tracking and detection. Code is publicly available.
Leveraging Uncertainty for Deep Interpretable Classification and Weakly-Supervised Segmentation of Histology Images
May 12, 2022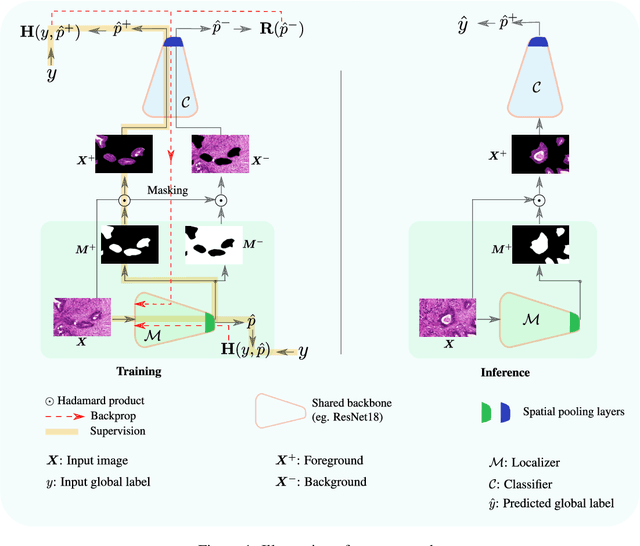
Abstract:Trained using only image class label, deep weakly supervised methods allow image classification and ROI segmentation for interpretability. Despite their success on natural images, they face several challenges over histology data where ROI are visually similar to background making models vulnerable to high pixel-wise false positives. These methods lack mechanisms for modeling explicitly non-discriminative regions which raises false-positive rates. We propose novel regularization terms, which enable the model to seek both non-discriminative and discriminative regions, while discouraging unbalanced segmentations and using only image class label. Our method is composed of two networks: a localizer that yields segmentation mask, followed by a classifier. The training loss pushes the localizer to build a segmentation mask that holds most discrimiantive regions while simultaneously modeling background regions. Comprehensive experiments over two histology datasets showed the merits of our method in reducing false positives and accurately segmenting ROI.
Negative Evidence Matters in Interpretable Histology Image Classification
Jan 07, 2022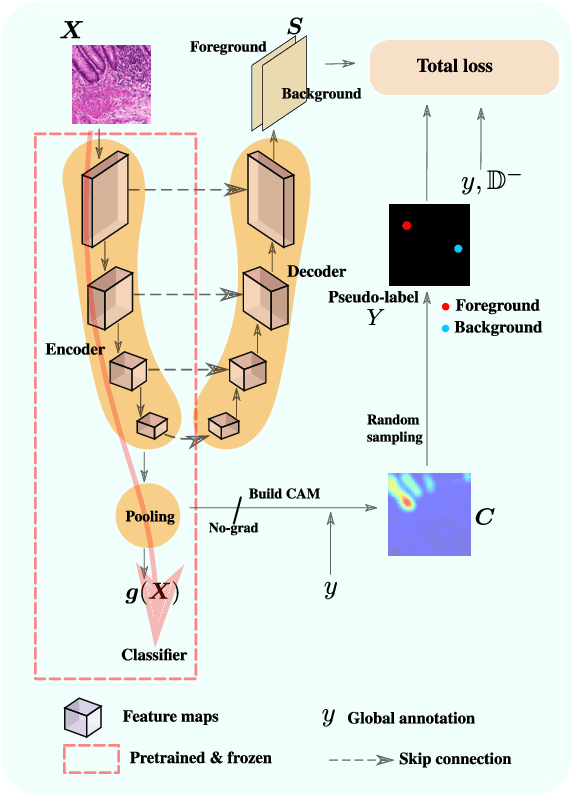
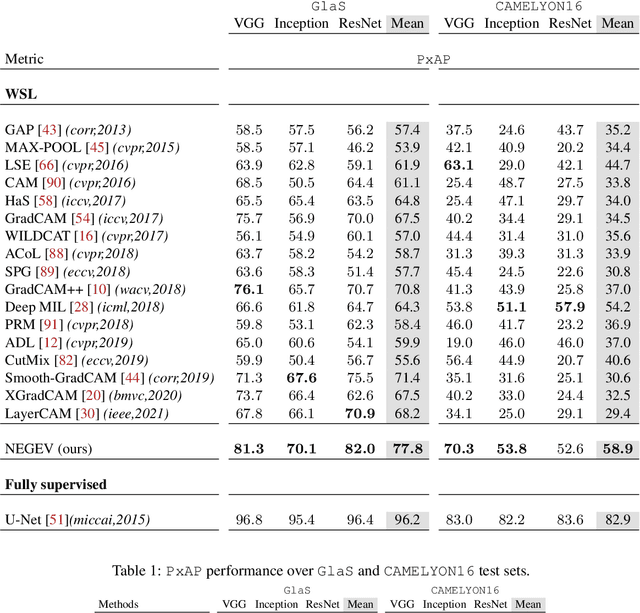
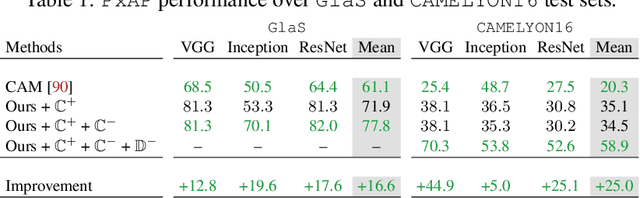
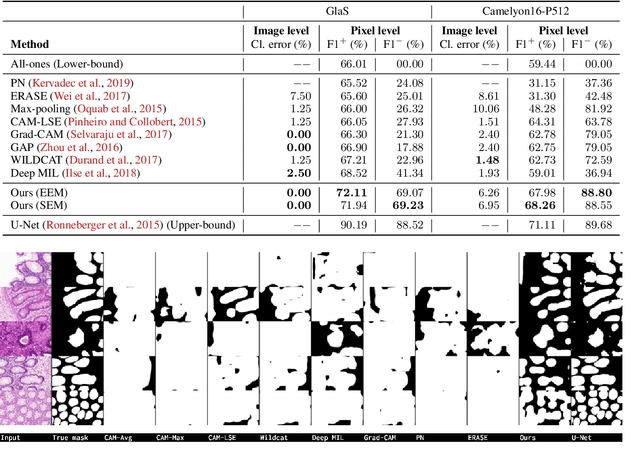
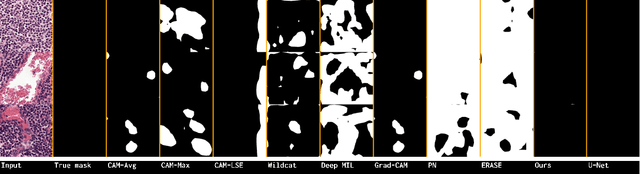
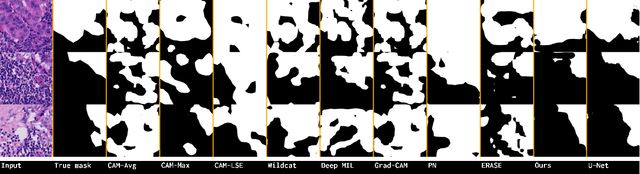
Abstract:Using only global annotations such as the image class labels, weakly-supervised learning methods allow CNN classifiers to jointly classify an image, and yield the regions of interest associated with the predicted class. However, without any guidance at the pixel level, such methods may yield inaccurate regions. This problem is known to be more challenging with histology images than with natural ones, since objects are less salient, structures have more variations, and foreground and background regions have stronger similarities. Therefore, methods in computer vision literature for visual interpretation of CNNs may not directly apply. In this work, we propose a simple yet efficient method based on a composite loss function that leverages information from the fully negative samples. Our new loss function contains two complementary terms: the first exploits positive evidence collected from the CNN classifier, while the second leverages the fully negative samples from the training dataset. In particular, we equip a pre-trained classifier with a decoder that allows refining the regions of interest. The same classifier is exploited to collect both the positive and negative evidence at the pixel level to train the decoder. This enables to take advantages of the fully negative samples that occurs naturally in the data, without any additional supervision signals and using only the image class as supervision. Compared to several recent related methods, over the public benchmark GlaS for colon cancer and a Camelyon16 patch-based benchmark for breast cancer using three different backbones, we show the substantial improvements introduced by our method. Our results shows the benefits of using both negative and positive evidence, ie, the one obtained from a classifier and the one naturally available in datasets. We provide an ablation study of both terms. Our code is publicly available.
F-CAM: Full Resolution CAM via Guided Parametric Upscaling
Sep 15, 2021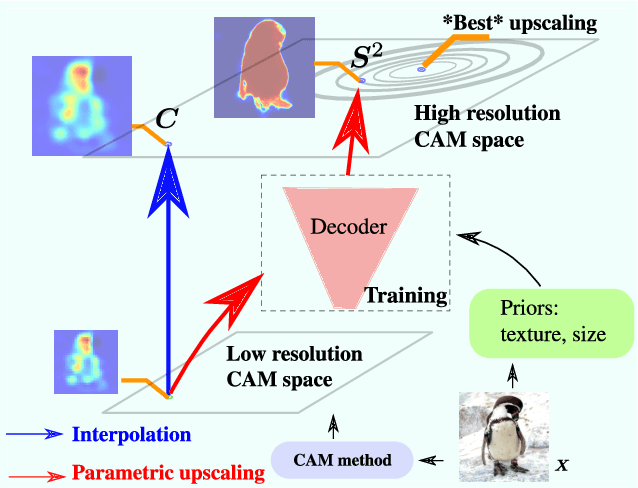
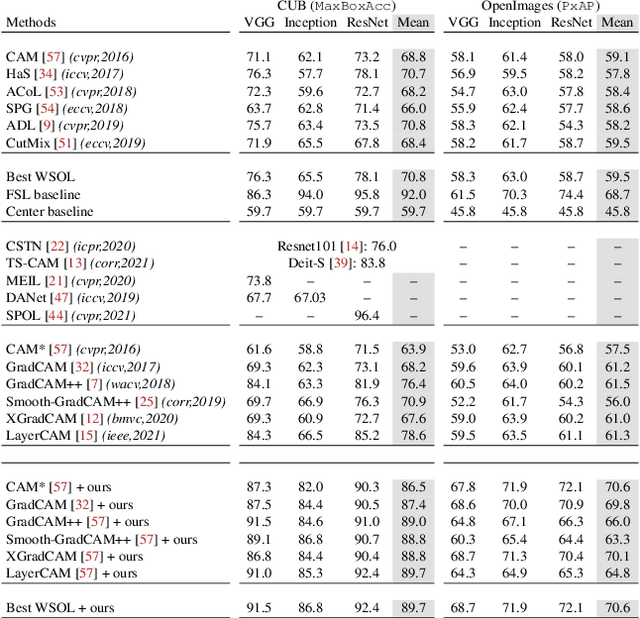
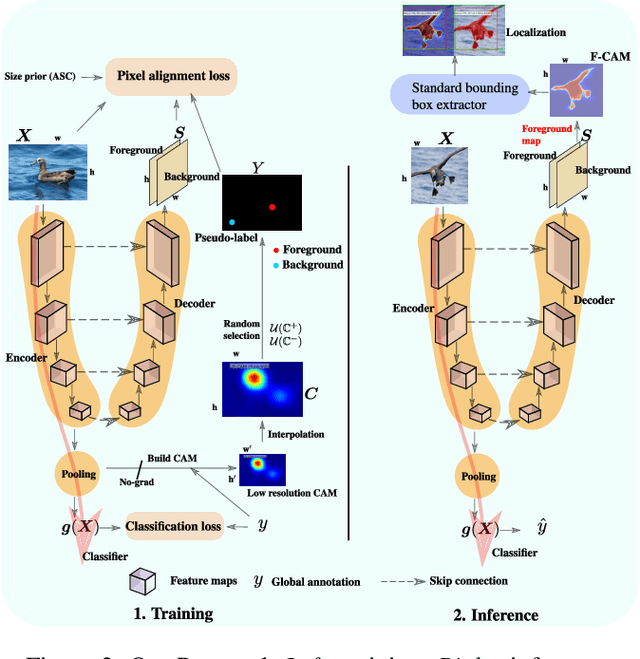
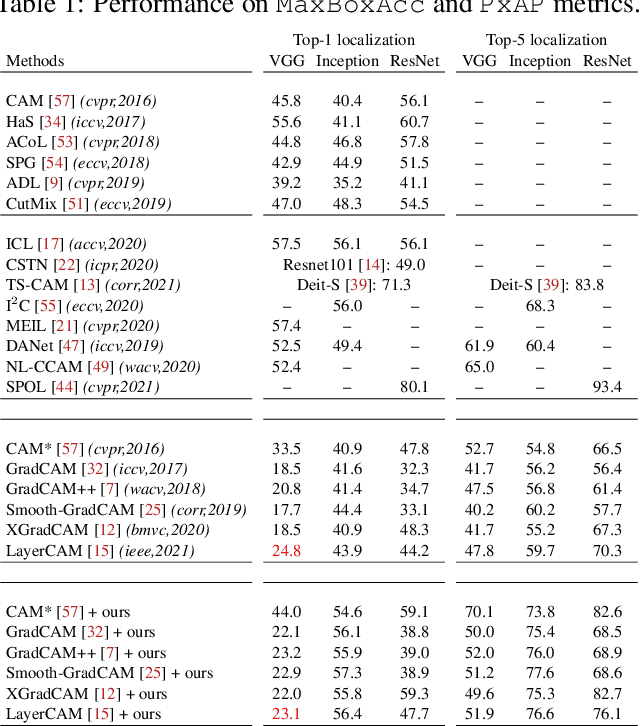
Abstract:Class Activation Mapping (CAM) methods have recently gained much attention for weakly-supervised object localization (WSOL) tasks, allowing for CNN visualization and interpretation without training on fully annotated image datasets. CAM methods are typically integrated within off-the-shelf CNN backbones, such as ResNet50. Due to convolution and downsampling/pooling operations, these backbones yield low resolution CAMs with a down-scaling factor of up to 32, making accurate localization more difficult. Interpolation is required to restore a full size CAMs, but without considering the statistical properties of the objects, leading to activations with inconsistent boundaries and inaccurate localizations. As an alternative, we introduce a generic method for parametric upscaling of CAMs that allows constructing accurate full resolution CAMs (F-CAMs). In particular, we propose a trainable decoding architecture that can be connected to any CNN classifier to produce more accurate CAMs. Given an original (low resolution) CAM, foreground and background pixels are randomly sampled for fine-tuning the decoder. Additional priors such as image statistics, and size constraints are also considered to expand and refine object boundaries. Extensive experiments using three CNN backbones and six WSOL baselines on the CUB-200-2011 and OpenImages datasets, indicate that our F-CAM method yields a significant improvement in CAM localization accuracy. F-CAM performance is competitive with state-of-art WSOL methods, yet it requires fewer computational resources during inference.
Deep Interpretable Classification and Weakly-Supervised Segmentation of Histology Images via Max-Min Uncertainty
Nov 14, 2020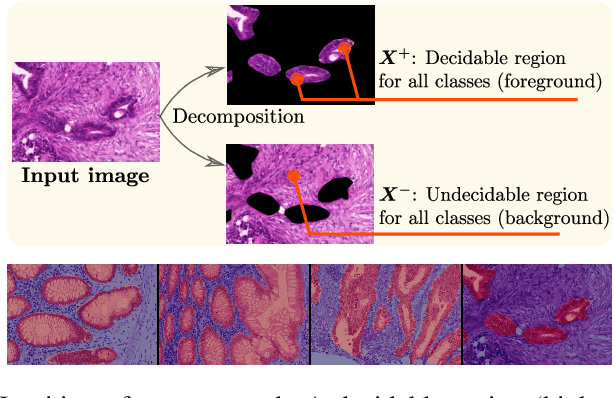
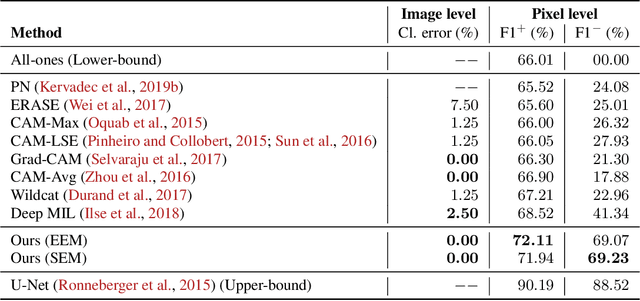
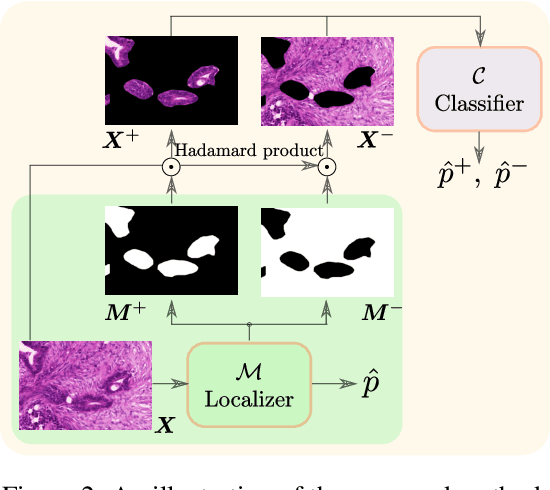
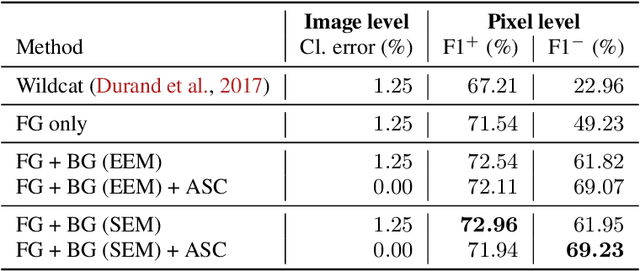
Abstract:Weakly supervised learning (WSL) has recently triggered substantial interest as it mitigates the lack of pixel-wise annotations, while enabling interpretable models. Given global image labels, WSL methods yield pixel-level predictions (segmentations). Despite their recent success, mostly with natural images, such methods could be seriously challenged when the foreground and background regions have similar visual cues, yielding high false-positive rates in segmentations, as is the case of challenging histology images. WSL training is commonly driven by standard classification losses, which implicitly maximize model confidence and find the discriminative regions linked to classification decisions. Therefore, they lack mechanisms for modeling explicitly non-discriminative regions and reducing false-positive rates. We propose new regularization terms, which enable the model to seek both non-discriminative and discriminative regions, while discouraging unbalanced segmentations. We introduce high uncertainty as a criterion to localize non-discriminative regions that do not affect classifier decision, and describe it with original Kullback-Leibler (KL) divergence losses evaluating the deviation of posterior predictions from the uniform distribution. Our KL terms encourage high uncertainty of the model when the latter takes the latent non-discriminative regions as input. Our loss integrates: (i) a cross-entropy seeking a foreground, where model confidence about class prediction is high; (ii) a KL regularizer seeking a background, where model uncertainty is high; and (iii) log-barrier terms discouraging unbalanced segmentations. Comprehensive experiments and ablation studies over the public GlaS colon cancer data show substantial improvements over state-of-the-art WSL methods, and confirm the effect of our new regularizers. Our code is publicly available.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge