Negative Evidence Matters in Interpretable Histology Image Classification
Paper and Code
Jan 07, 2022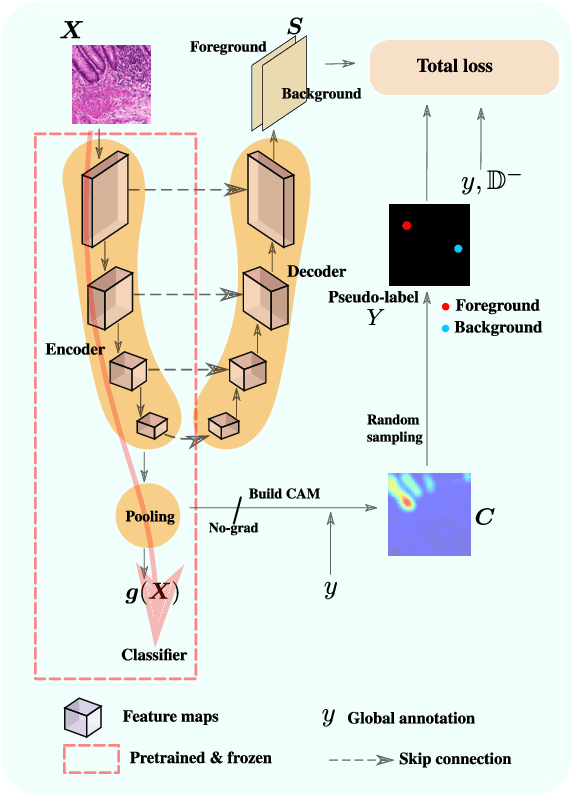
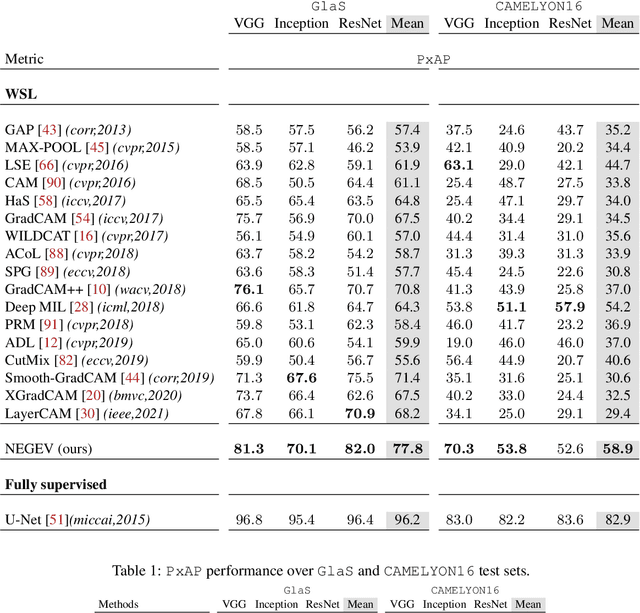
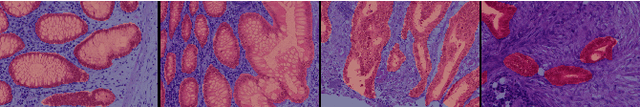
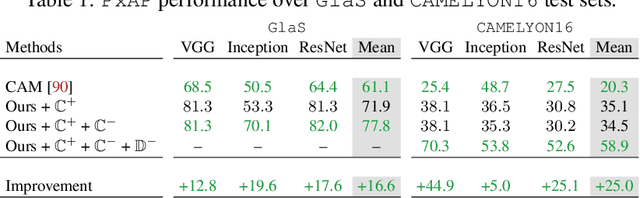
Using only global annotations such as the image class labels, weakly-supervised learning methods allow CNN classifiers to jointly classify an image, and yield the regions of interest associated with the predicted class. However, without any guidance at the pixel level, such methods may yield inaccurate regions. This problem is known to be more challenging with histology images than with natural ones, since objects are less salient, structures have more variations, and foreground and background regions have stronger similarities. Therefore, methods in computer vision literature for visual interpretation of CNNs may not directly apply. In this work, we propose a simple yet efficient method based on a composite loss function that leverages information from the fully negative samples. Our new loss function contains two complementary terms: the first exploits positive evidence collected from the CNN classifier, while the second leverages the fully negative samples from the training dataset. In particular, we equip a pre-trained classifier with a decoder that allows refining the regions of interest. The same classifier is exploited to collect both the positive and negative evidence at the pixel level to train the decoder. This enables to take advantages of the fully negative samples that occurs naturally in the data, without any additional supervision signals and using only the image class as supervision. Compared to several recent related methods, over the public benchmark GlaS for colon cancer and a Camelyon16 patch-based benchmark for breast cancer using three different backbones, we show the substantial improvements introduced by our method. Our results shows the benefits of using both negative and positive evidence, ie, the one obtained from a classifier and the one naturally available in datasets. We provide an ablation study of both terms. Our code is publicly available.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge