Vincent Morard
Multi-Task Multi-Scale Learning For Outcome Prediction in 3D PET Images
Mar 01, 2022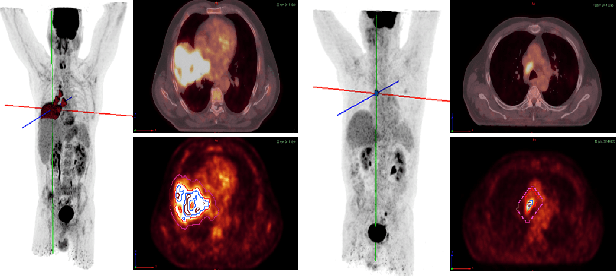
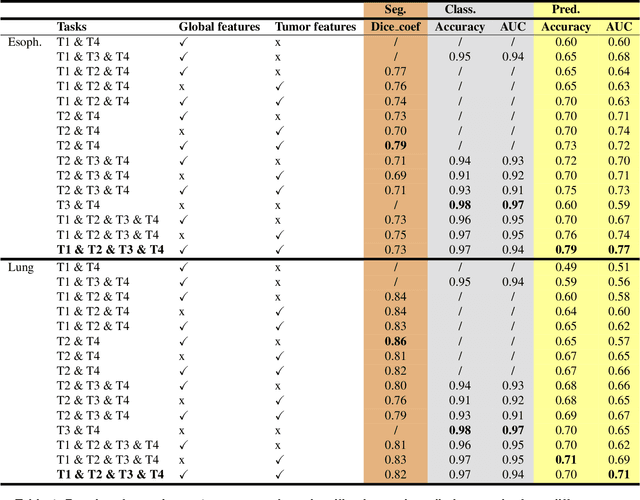
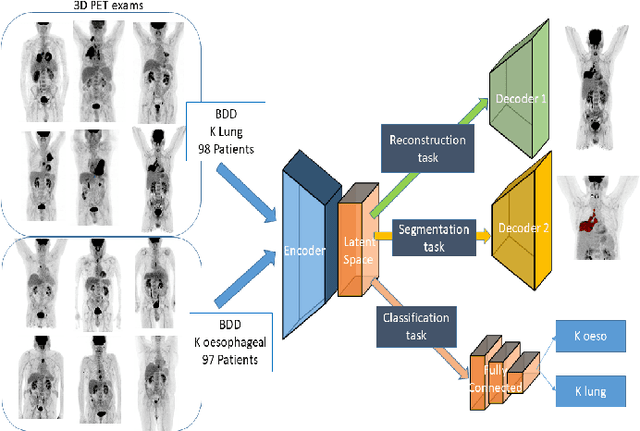
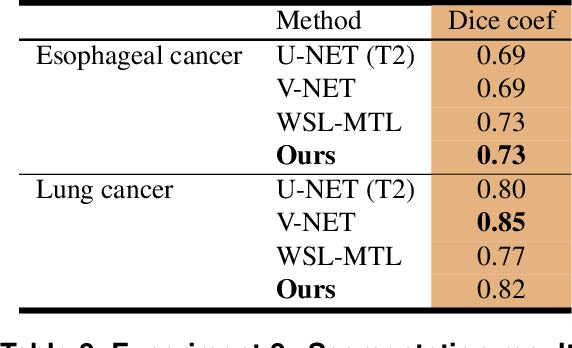
Abstract:Background and Objectives: Predicting patient response to treatment and survival in oncology is a prominent way towards precision medicine. To that end, radiomics was proposed as a field of study where images are used instead of invasive methods. The first step in radiomic analysis is the segmentation of the lesion. However, this task is time consuming and can be physician subjective. Automated tools based on supervised deep learning have made great progress to assist physicians. However, they are data hungry, and annotated data remains a major issue in the medical field where only a small subset of annotated images is available. Methods: In this work, we propose a multi-task learning framework to predict patient's survival and response. We show that the encoder can leverage multiple tasks to extract meaningful and powerful features that improve radiomics performance. We show also that subsidiary tasks serve as an inductive bias so that the model can better generalize. Results: Our model was tested and validated for treatment response and survival in lung and esophageal cancers, with an area under the ROC curve of 77% and 71% respectively, outperforming single task learning methods. Conclusions: We show that, by using a multi-task learning approach, we can boost the performance of radiomic analysis by extracting rich information of intratumoral and peritumoral regions.
Weakly Supervised PET Tumor Detection Using Class Response
Mar 19, 2020



Abstract:One of the most challenges in medical imaging is the lack of data and annotated data. It is proven that classical segmentation methods such as U-NET are useful but still limited due to the lack of annotated data. Using a weakly supervised learning is a promising way to address this problem, however, it is challenging to train one model to detect and locate efficiently different type of lesions due to the huge variation in images. In this paper, we present a novel approach to locate different type of lesions in positron emission tomography (PET) images using only a class label at the image-level. First, a simple convolutional neural network classifier is trained to predict the type of cancer on two 2D MIP images. Then, a pseudo-localization of the tumor is generated using class activation maps, back-propagated and corrected in a multitask learning approach with prior knowledge, resulting in a tumor detection mask. Finally, we use the mask generated from the two 2D images to detect the tumor in the 3D image. The advantage of our proposed method consists of detecting the whole tumor volume in 3D images, using only two 2D images of PET image, and showing a very promising results. It can be used as a tool to locate very efficiently tumors in a PET scan, which is a time-consuming task for physicians. In addition, we show that our proposed method can be used to conduct a radiomics study with state of the art results.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge