Pierre Vera
Mamba Based Feature Extraction And Adaptive Multilevel Feature Fusion For 3D Tumor Segmentation From Multi-modal Medical Image
Apr 30, 2025Abstract:Multi-modal 3D medical image segmentation aims to accurately identify tumor regions across different modalities, facing challenges from variations in image intensity and tumor morphology. Traditional convolutional neural network (CNN)-based methods struggle with capturing global features, while Transformers-based methods, despite effectively capturing global context, encounter high computational costs in 3D medical image segmentation. The Mamba model combines linear scalability with long-distance modeling, making it a promising approach for visual representation learning. However, Mamba-based 3D multi-modal segmentation still struggles to leverage modality-specific features and fuse complementary information effectively. In this paper, we propose a Mamba based feature extraction and adaptive multilevel feature fusion for 3D tumor segmentation using multi-modal medical image. We first develop the specific modality Mamba encoder to efficiently extract long-range relevant features that represent anatomical and pathological structures present in each modality. Moreover, we design an bi-level synergistic integration block that dynamically merges multi-modal and multi-level complementary features by the modality attention and channel attention learning. Lastly, the decoder combines deep semantic information with fine-grained details to generate the tumor segmentation map. Experimental results on medical image datasets (PET/CT and MRI multi-sequence) show that our approach achieve competitive performance compared to the state-of-the-art CNN, Transformer, and Mamba-based approaches.
Iterative Collaboration Network Guided By Reconstruction Prior for Medical Image Super-Resolution
Apr 23, 2025Abstract:High-resolution medical images can provide more detailed information for better diagnosis. Conventional medical image super-resolution relies on a single task which first performs the extraction of the features and then upscaling based on the features. The features extracted may not be complete for super-resolution. Recent multi-task learning,including reconstruction and super-resolution, is a good solution to obtain additional relevant information. The interaction between the two tasks is often insufficient, which still leads to incomplete and less relevant deep features. To address above limitations, we propose an iterative collaboration network (ICONet) to improve communications between tasks by progressively incorporating reconstruction prior to the super-resolution learning procedure in an iterative collaboration way. It consists of a reconstruction branch, a super-resolution branch, and a SR-Rec fusion module. The reconstruction branch generates the artifact-free image as prior, which is followed by a super-resolution branch for prior knowledge-guided super-resolution. Unlike the widely-used convolutional neural networks for extracting local features and Transformers with quadratic computational complexity for modeling long-range dependencies, we develop a new residual spatial-channel feature learning (RSCFL) module of two branches to efficiently establish feature relationships in spatial and channel dimensions. Moreover, the designed SR-Rec fusion module fuses the reconstruction prior and super-resolution features with each other in an adaptive manner. Our ICONet is built with multi-stage models to iteratively upscale the low-resolution images using steps of 2x and simultaneously interact between two branches in multi-stage supervisions.
Deform-Mamba Network for MRI Super-Resolution
Jul 08, 2024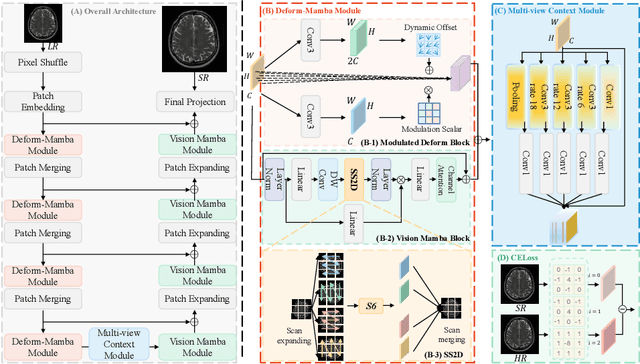
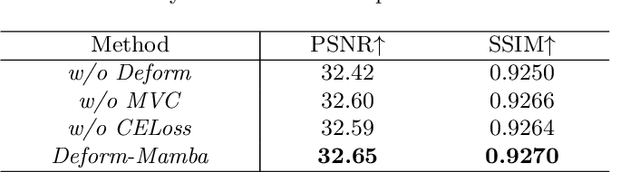
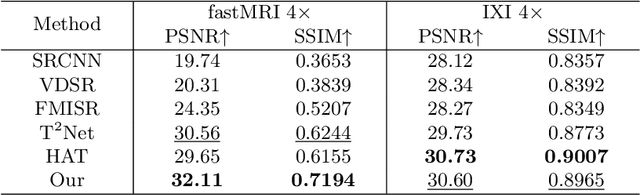
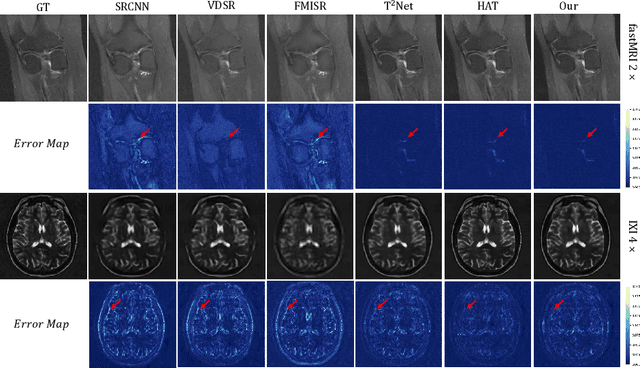
Abstract:In this paper, we propose a new architecture, called Deform-Mamba, for MR image super-resolution. Unlike conventional CNN or Transformer-based super-resolution approaches which encounter challenges related to the local respective field or heavy computational cost, our approach aims to effectively explore the local and global information of images. Specifically, we develop a Deform-Mamba encoder which is composed of two branches, modulated deform block and vision Mamba block. We also design a multi-view context module in the bottleneck layer to explore the multi-view contextual content. Thanks to the extracted features of the encoder, which include content-adaptive local and efficient global information, the vision Mamba decoder finally generates high-quality MR images. Moreover, we introduce a contrastive edge loss to promote the reconstruction of edge and contrast related content. Quantitative and qualitative experimental results indicate that our approach on IXI and fastMRI datasets achieves competitive performance.
Self-Prior Guided Mamba-UNet Networks for Medical Image Super-Resolution
Jul 08, 2024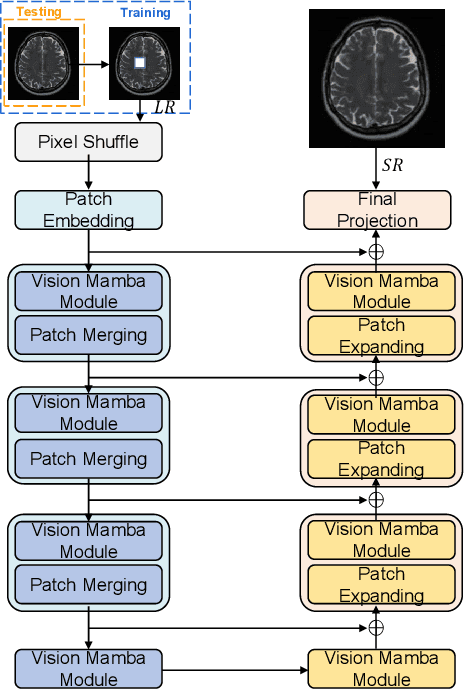
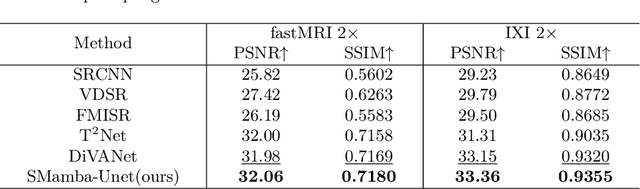
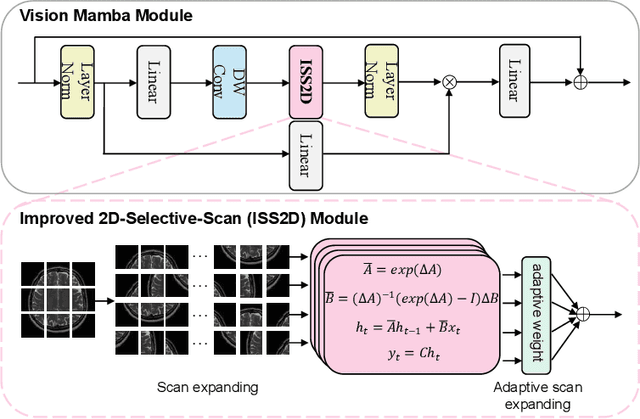
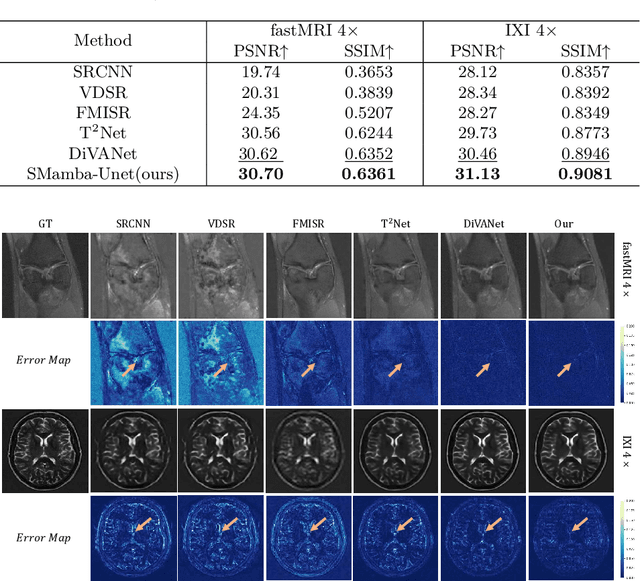
Abstract:In this paper, we propose a self-prior guided Mamba-UNet network (SMamba-UNet) for medical image super-resolution. Existing methods are primarily based on convolutional neural networks (CNNs) or Transformers. CNNs-based methods fail to capture long-range dependencies, while Transformer-based approaches face heavy calculation challenges due to their quadratic computational complexity. Recently, State Space Models (SSMs) especially Mamba have emerged, capable of modeling long-range dependencies with linear computational complexity. Inspired by Mamba, our approach aims to learn the self-prior multi-scale contextual features under Mamba-UNet networks, which may help to super-resolve low-resolution medical images in an efficient way. Specifically, we obtain self-priors by perturbing the brightness inpainting of the input image during network training, which can learn detailed texture and brightness information that is beneficial for super-resolution. Furthermore, we combine Mamba with Unet network to mine global features at different levels. We also design an improved 2D-Selective-Scan (ISS2D) module to divide image features into different directional sequences to learn long-range dependencies in multiple directions, and adaptively fuse sequence information to enhance super-resolved feature representation. Both qualitative and quantitative experimental results demonstrate that our approach outperforms current state-of-the-art methods on two public medical datasets: the IXI and fastMRI.
Discriminative Hamiltonian Variational Autoencoder for Accurate Tumor Segmentation in Data-Scarce Regimes
Jun 17, 2024



Abstract:Deep learning has gained significant attention in medical image segmentation. However, the limited availability of annotated training data presents a challenge to achieving accurate results. In efforts to overcome this challenge, data augmentation techniques have been proposed. However, the majority of these approaches primarily focus on image generation. For segmentation tasks, providing both images and their corresponding target masks is crucial, and the generation of diverse and realistic samples remains a complex task, especially when working with limited training datasets. To this end, we propose a new end-to-end hybrid architecture based on Hamiltonian Variational Autoencoders (HVAE) and a discriminative regularization to improve the quality of generated images. Our method provides an accuracte estimation of the joint distribution of the images and masks, resulting in the generation of realistic medical images with reduced artifacts and off-distribution instances. As generating 3D volumes requires substantial time and memory, our architecture operates on a slice-by-slice basis to segment 3D volumes, capitilizing on the richly augmented dataset. Experiments conducted on two public datasets, BRATS (MRI modality) and HECKTOR (PET modality), demonstrate the efficacy of our proposed method on different medical imaging modalities with limited data.
3D MRI Synthesis with Slice-Based Latent Diffusion Models: Improving Tumor Segmentation Tasks in Data-Scarce Regimes
Jun 08, 2024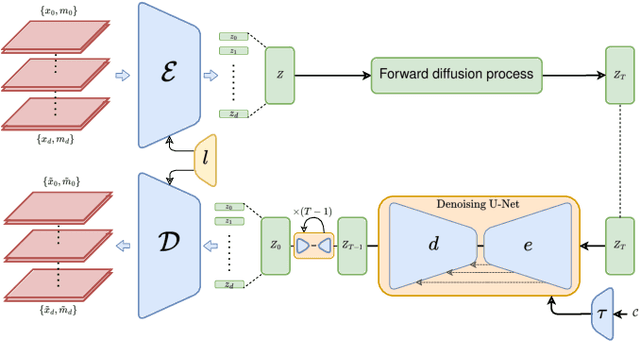
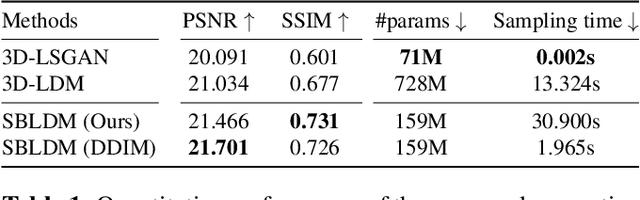
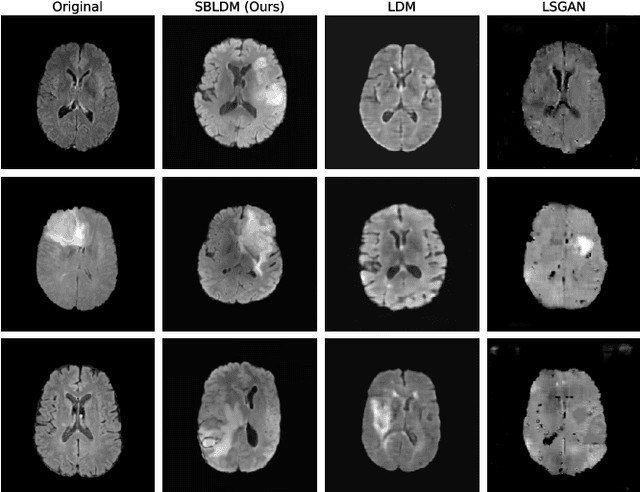
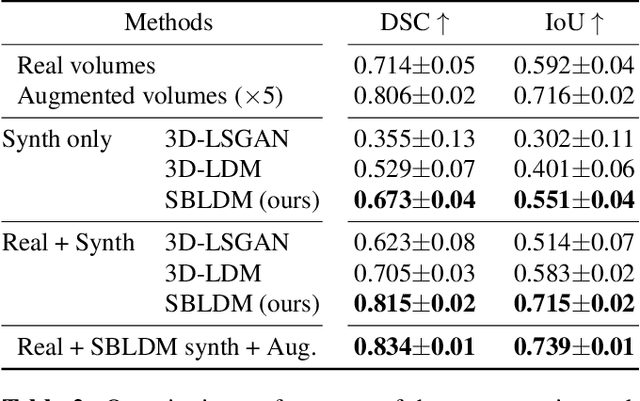
Abstract:Despite the increasing use of deep learning in medical image segmentation, the limited availability of annotated training data remains a major challenge due to the time-consuming data acquisition and privacy regulations. In the context of segmentation tasks, providing both medical images and their corresponding target masks is essential. However, conventional data augmentation approaches mainly focus on image synthesis. In this study, we propose a novel slice-based latent diffusion architecture designed to address the complexities of volumetric data generation in a slice-by-slice fashion. This approach extends the joint distribution modeling of medical images and their associated masks, allowing a simultaneous generation of both under data-scarce regimes. Our approach mitigates the computational complexity and memory expensiveness typically associated with diffusion models. Furthermore, our architecture can be conditioned by tumor characteristics, including size, shape, and relative position, thereby providing a diverse range of tumor variations. Experiments on a segmentation task using the BRATS2022 confirm the effectiveness of the synthesized volumes and masks for data augmentation.
End-to-end autoencoding architecture for the simultaneous generation of medical images and corresponding segmentation masks
Nov 17, 2023Abstract:Despite the increasing use of deep learning in medical image segmentation, acquiring sufficient training data remains a challenge in the medical field. In response, data augmentation techniques have been proposed; however, the generation of diverse and realistic medical images and their corresponding masks remains a difficult task, especially when working with insufficient training sets. To address these limitations, we present an end-to-end architecture based on the Hamiltonian Variational Autoencoder (HVAE). This approach yields an improved posterior distribution approximation compared to traditional Variational Autoencoders (VAE), resulting in higher image generation quality. Our method outperforms generative adversarial architectures under data-scarce conditions, showcasing enhancements in image quality and precise tumor mask synthesis. We conduct experiments on two publicly available datasets, MICCAI's Brain Tumor Segmentation Challenge (BRATS), and Head and Neck Tumor Segmentation Challenge (HECKTOR), demonstrating the effectiveness of our method on different medical imaging modalities.
Evidence fusion with contextual discounting for multi-modality medical image segmentation
Jun 27, 2022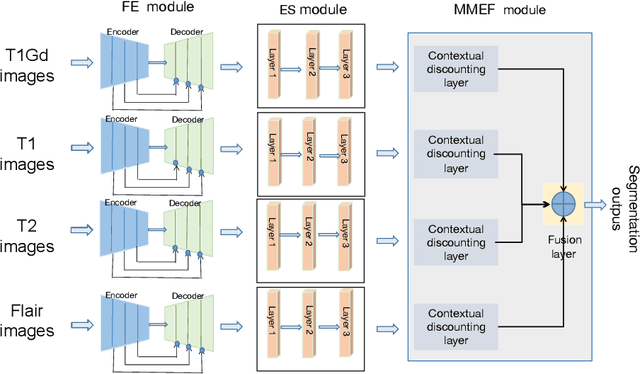
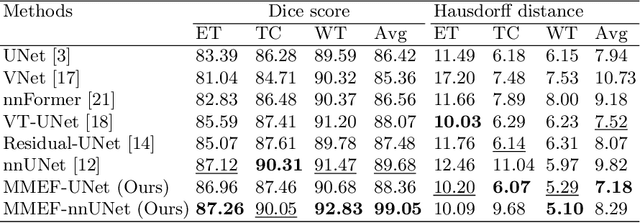
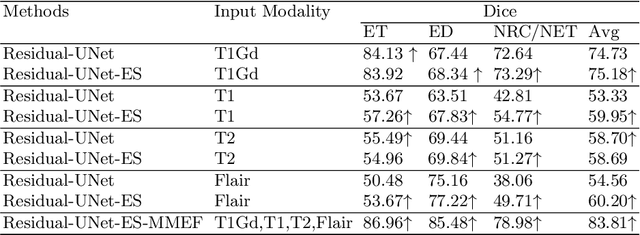
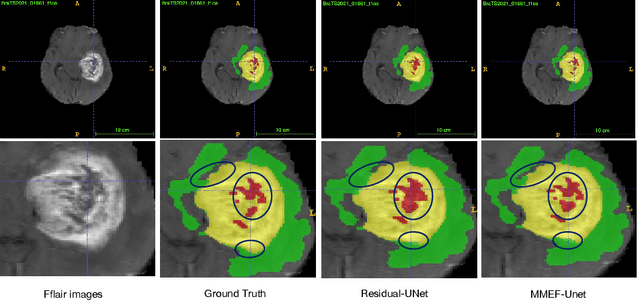
Abstract:As information sources are usually imperfect, it is necessary to take into account their reliability in multi-source information fusion tasks. In this paper, we propose a new deep framework allowing us to merge multi-MR image segmentation results using the formalism of Dempster-Shafer theory while taking into account the reliability of different modalities relative to different classes. The framework is composed of an encoder-decoder feature extraction module, an evidential segmentation module that computes a belief function at each voxel for each modality, and a multi-modality evidence fusion module, which assigns a vector of discount rates to each modality evidence and combines the discounted evidence using Dempster's rule. The whole framework is trained by minimizing a new loss function based on a discounted Dice index to increase segmentation accuracy and reliability. The method was evaluated on the BraTs 2021 database of 1251 patients with brain tumors. Quantitative and qualitative results show that our method outperforms the state of the art, and implements an effective new idea for merging multi-information within deep neural networks.
A Quantitative Comparison between Shannon and Tsallis Havrda Charvat Entropies Applied to Cancer Outcome Prediction
Mar 22, 2022
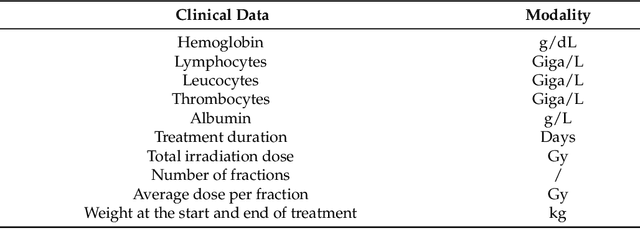
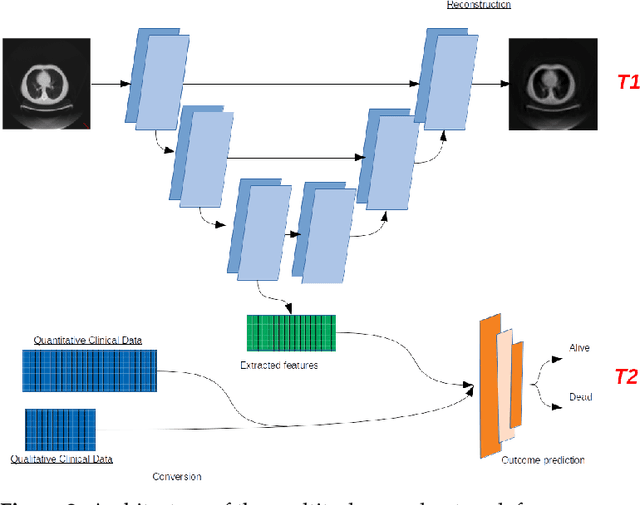

Abstract:In this paper, we propose to quantitatively compare loss functions based on parameterized Tsallis-Havrda-Charvat entropy and classical Shannon entropy for the training of a deep network in the case of small datasets which are usually encountered in medical applications. Shannon cross-entropy is widely used as a loss function for most neural networks applied to the segmentation, classification and detection of images. Shannon entropy is a particular case of Tsallis-Havrda-Charvat entropy. In this work, we compare these two entropies through a medical application for predicting recurrence in patients with head-neck and lung cancers after treatment. Based on both CT images and patient information, a multitask deep neural network is proposed to perform a recurrence prediction task using cross-entropy as a loss function and an image reconstruction task. Tsallis-Havrda-Charvat cross-entropy is a parameterized cross entropy with the parameter $\alpha$. Shannon entropy is a particular case of Tsallis-Havrda-Charvat entropy for $\alpha$ = 1. The influence of this parameter on the final prediction results is studied. In this paper, the experiments are conducted on two datasets including in total 580 patients, of whom 434 suffered from head-neck cancers and 146 from lung cancers. The results show that Tsallis-Havrda-Charvat entropy can achieve better performance in terms of prediction accuracy with some values of $\alpha$.
* 11 pages, 3 figures
Multi-Task Multi-Scale Learning For Outcome Prediction in 3D PET Images
Mar 01, 2022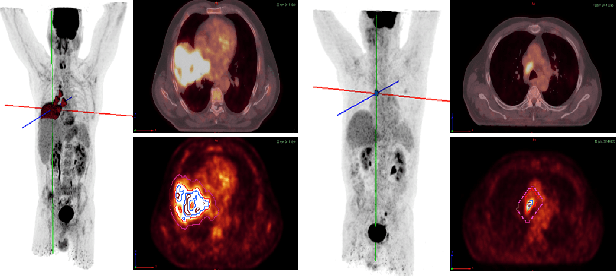
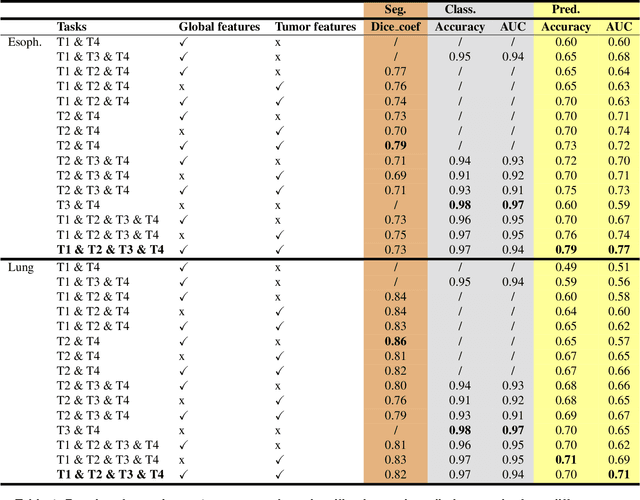
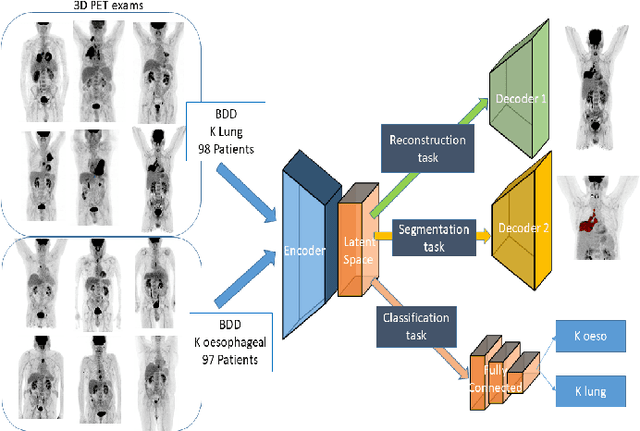
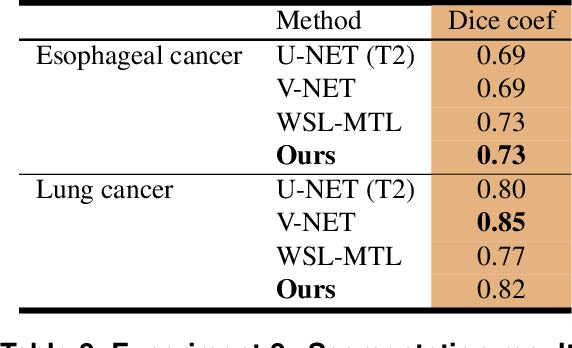
Abstract:Background and Objectives: Predicting patient response to treatment and survival in oncology is a prominent way towards precision medicine. To that end, radiomics was proposed as a field of study where images are used instead of invasive methods. The first step in radiomic analysis is the segmentation of the lesion. However, this task is time consuming and can be physician subjective. Automated tools based on supervised deep learning have made great progress to assist physicians. However, they are data hungry, and annotated data remains a major issue in the medical field where only a small subset of annotated images is available. Methods: In this work, we propose a multi-task learning framework to predict patient's survival and response. We show that the encoder can leverage multiple tasks to extract meaningful and powerful features that improve radiomics performance. We show also that subsidiary tasks serve as an inductive bias so that the model can better generalize. Results: Our model was tested and validated for treatment response and survival in lung and esophageal cancers, with an area under the ROC curve of 77% and 71% respectively, outperforming single task learning methods. Conclusions: We show that, by using a multi-task learning approach, we can boost the performance of radiomic analysis by extracting rich information of intratumoral and peritumoral regions.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge