Qingchun Liang
SMILE: a Scale-aware Multiple Instance Learning Method for Multicenter STAS Lung Cancer Histopathology Diagnosis
Mar 18, 2025Abstract:Spread through air spaces (STAS) represents a newly identified aggressive pattern in lung cancer, which is known to be associated with adverse prognostic factors and complex pathological features. Pathologists currently rely on time consuming manual assessments, which are highly subjective and prone to variation. This highlights the urgent need for automated and precise diag nostic solutions. 2,970 lung cancer tissue slides are comprised from multiple centers, re-diagnosed them, and constructed and publicly released three lung cancer STAS datasets: STAS CSU (hospital), STAS TCGA, and STAS CPTAC. All STAS datasets provide corresponding pathological feature diagnoses and related clinical data. To address the bias, sparse and heterogeneous nature of STAS, we propose an scale-aware multiple instance learning(SMILE) method for STAS diagnosis of lung cancer. By introducing a scale-adaptive attention mechanism, the SMILE can adaptively adjust high attention instances, reducing over-reliance on local regions and promoting consistent detection of STAS lesions. Extensive experiments show that SMILE achieved competitive diagnostic results on STAS CSU, diagnosing 251 and 319 STAS samples in CPTAC andTCGA,respectively, surpassing clinical average AUC. The 11 open baseline results are the first to be established for STAS research, laying the foundation for the future expansion, interpretability, and clinical integration of computational pathology technologies. The datasets and code are available at https://anonymous.4open.science/r/IJCAI25-1DA1.
Feature-interactive Siamese graph encoder-based image analysis to predict STAS from histopathology images in lung cancer
Nov 22, 2024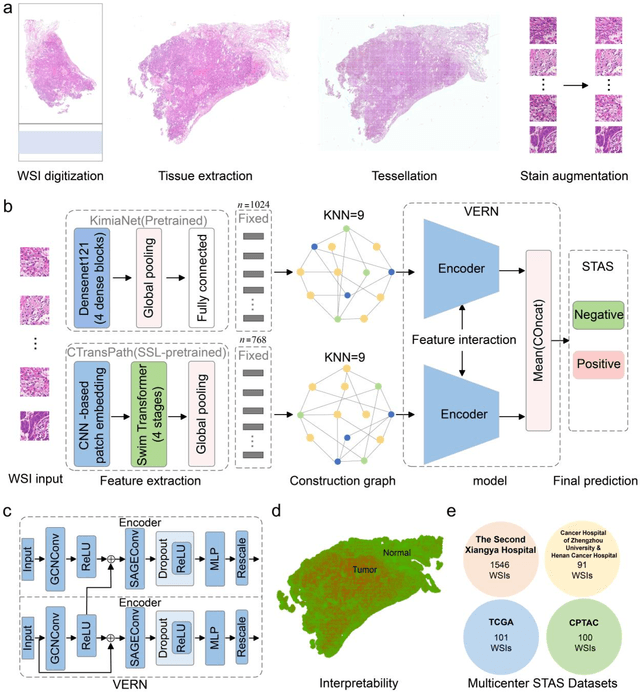
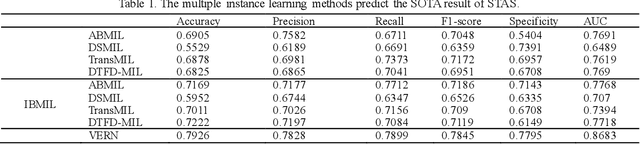
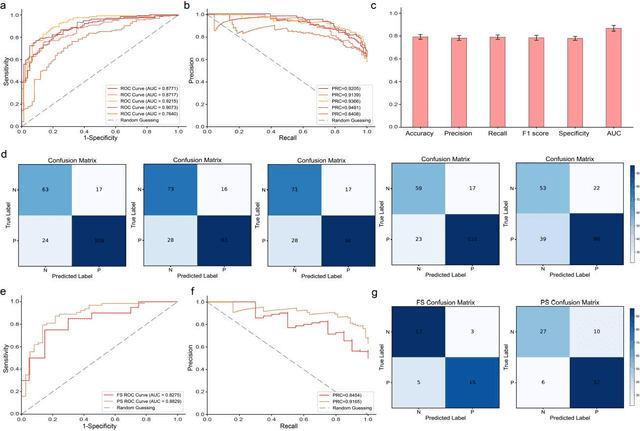
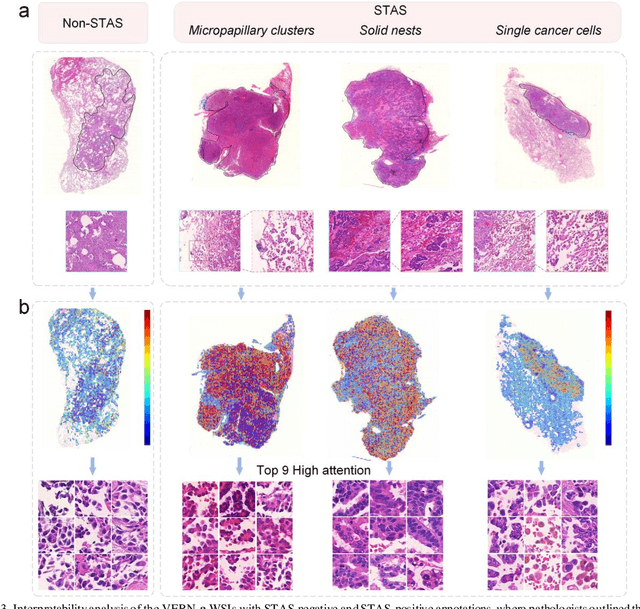
Abstract:Spread through air spaces (STAS) is a distinct invasion pattern in lung cancer, crucial for prognosis assessment and guiding surgical decisions. Histopathology is the gold standard for STAS detection, yet traditional methods are subjective, time-consuming, and prone to misdiagnosis, limiting large-scale applications. We present VERN, an image analysis model utilizing a feature-interactive Siamese graph encoder to predict STAS from lung cancer histopathological images. VERN captures spatial topological features with feature sharing and skip connections to enhance model training. Using 1,546 histopathology slides, we built a large single-cohort STAS lung cancer dataset. VERN achieved an AUC of 0.9215 in internal validation and AUCs of 0.8275 and 0.8829 in frozen and paraffin-embedded test sections, respectively, demonstrating clinical-grade performance. Validated on a single-cohort and three external datasets, VERN showed robust predictive performance and generalizability, providing an open platform (http://plr.20210706.xyz:5000/) to enhance STAS diagnosis efficiency and accuracy.
FORESEE: Multimodal and Multi-view Representation Learning for Robust Prediction of Cancer Survival
May 13, 2024Abstract:Integrating the different data modalities of cancer patients can significantly improve the predictive performance of patient survival. However, most existing methods ignore the simultaneous utilization of rich semantic features at different scales in pathology images. When collecting multimodal data and extracting features, there is a likelihood of encountering intra-modality missing data, introducing noise into the multimodal data. To address these challenges, this paper proposes a new end-to-end framework, FORESEE, for robustly predicting patient survival by mining multimodal information. Specifically, the cross-fusion transformer effectively utilizes features at the cellular level, tissue level, and tumor heterogeneity level to correlate prognosis through a cross-scale feature cross-fusion method. This enhances the ability of pathological image feature representation. Secondly, the hybrid attention encoder (HAE) uses the denoising contextual attention module to obtain the contextual relationship features and local detail features of the molecular data. HAE's channel attention module obtains global features of molecular data. Furthermore, to address the issue of missing information within modalities, we propose an asymmetrically masked triplet masked autoencoder to reconstruct lost information within modalities. Extensive experiments demonstrate the superiority of our method over state-of-the-art methods on four benchmark datasets in both complete and missing settings.
Opportunities and challenges in the application of large artificial intelligence models in radiology
Mar 24, 2024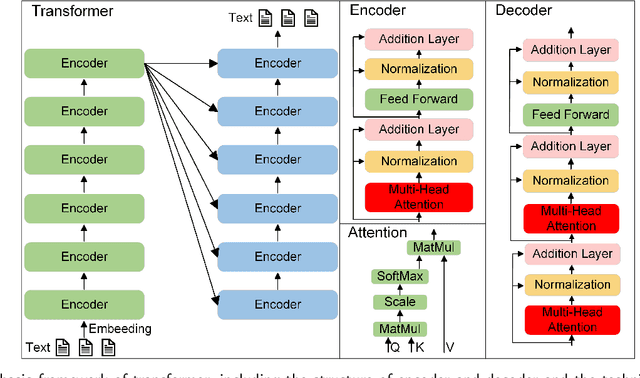
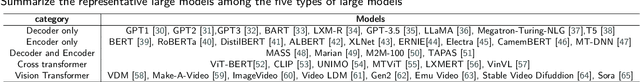
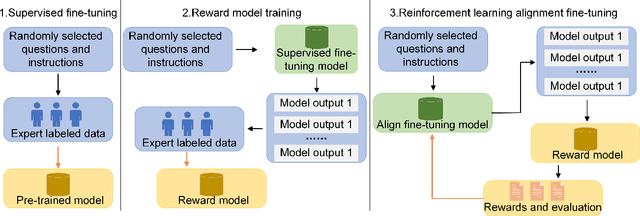
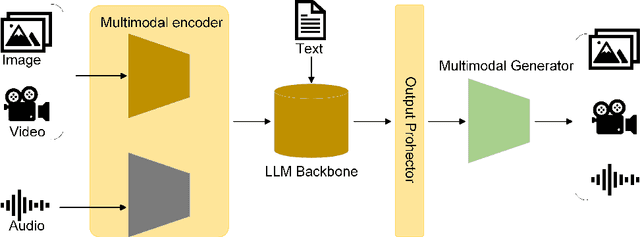
Abstract:Influenced by ChatGPT, artificial intelligence (AI) large models have witnessed a global upsurge in large model research and development. As people enjoy the convenience by this AI large model, more and more large models in subdivided fields are gradually being proposed, especially large models in radiology imaging field. This article first introduces the development history of large models, technical details, workflow, working principles of multimodal large models and working principles of video generation large models. Secondly, we summarize the latest research progress of AI large models in radiology education, radiology report generation, applications of unimodal and multimodal radiology. Finally, this paper also summarizes some of the challenges of large AI models in radiology, with the aim of better promoting the rapid revolution in the field of radiography.
SELECTOR: Heterogeneous graph network with convolutional masked autoencoder for multimodal robust prediction of cancer survival
Mar 14, 2024Abstract:Accurately predicting the survival rate of cancer patients is crucial for aiding clinicians in planning appropriate treatment, reducing cancer-related medical expenses, and significantly enhancing patients' quality of life. Multimodal prediction of cancer patient survival offers a more comprehensive and precise approach. However, existing methods still grapple with challenges related to missing multimodal data and information interaction within modalities. This paper introduces SELECTOR, a heterogeneous graph-aware network based on convolutional mask encoders for robust multimodal prediction of cancer patient survival. SELECTOR comprises feature edge reconstruction, convolutional mask encoder, feature cross-fusion, and multimodal survival prediction modules. Initially, we construct a multimodal heterogeneous graph and employ the meta-path method for feature edge reconstruction, ensuring comprehensive incorporation of feature information from graph edges and effective embedding of nodes. To mitigate the impact of missing features within the modality on prediction accuracy, we devised a convolutional masked autoencoder (CMAE) to process the heterogeneous graph post-feature reconstruction. Subsequently, the feature cross-fusion module facilitates communication between modalities, ensuring that output features encompass all features of the modality and relevant information from other modalities. Extensive experiments and analysis on six cancer datasets from TCGA demonstrate that our method significantly outperforms state-of-the-art methods in both modality-missing and intra-modality information-confirmed cases. Our codes are made available at https://github.com/panliangrui/Selector.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge