Wenwu Zeng
Feature-interactive Siamese graph encoder-based image analysis to predict STAS from histopathology images in lung cancer
Nov 22, 2024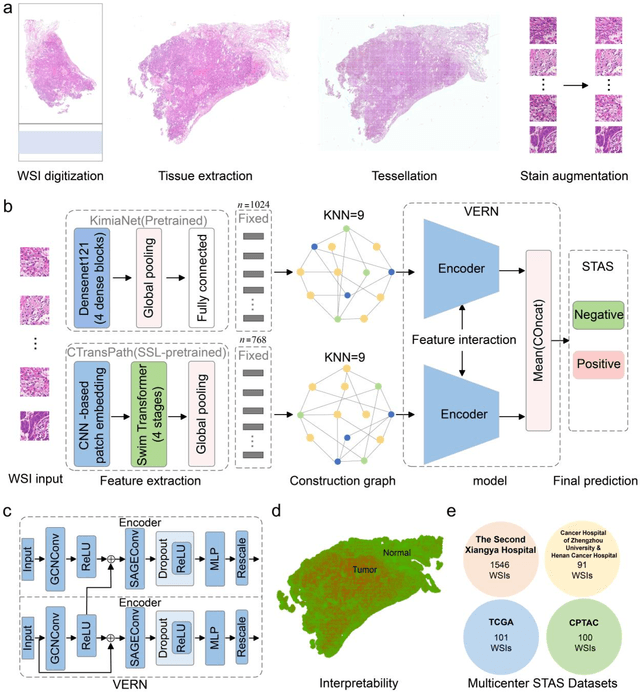
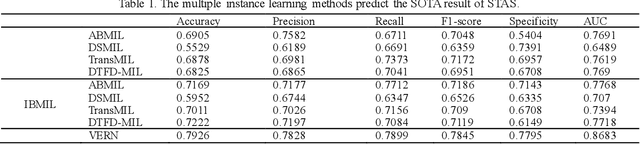
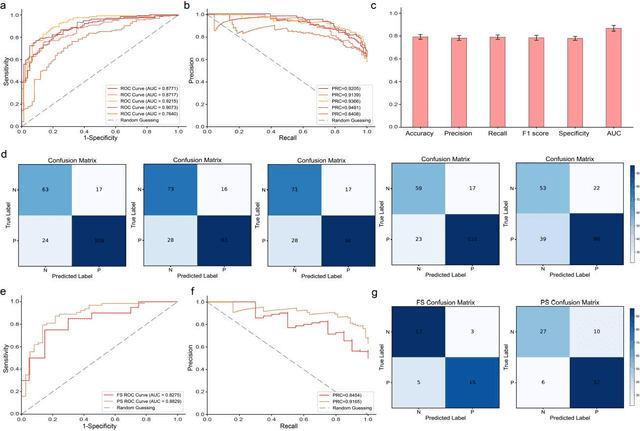
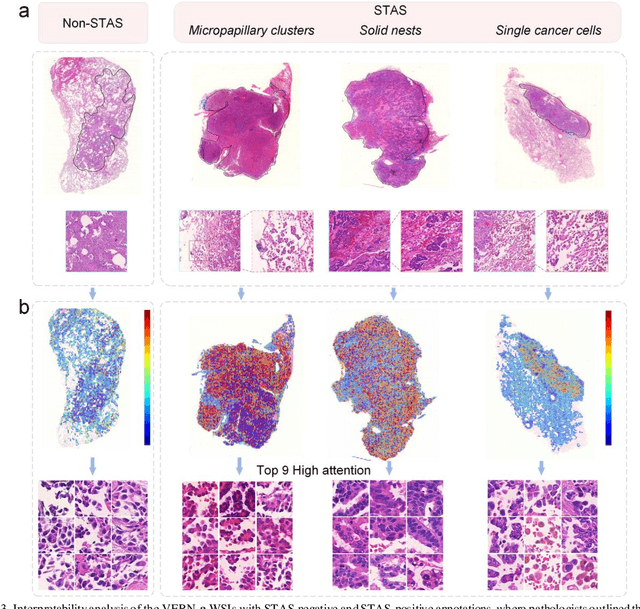
Abstract:Spread through air spaces (STAS) is a distinct invasion pattern in lung cancer, crucial for prognosis assessment and guiding surgical decisions. Histopathology is the gold standard for STAS detection, yet traditional methods are subjective, time-consuming, and prone to misdiagnosis, limiting large-scale applications. We present VERN, an image analysis model utilizing a feature-interactive Siamese graph encoder to predict STAS from lung cancer histopathological images. VERN captures spatial topological features with feature sharing and skip connections to enhance model training. Using 1,546 histopathology slides, we built a large single-cohort STAS lung cancer dataset. VERN achieved an AUC of 0.9215 in internal validation and AUCs of 0.8275 and 0.8829 in frozen and paraffin-embedded test sections, respectively, demonstrating clinical-grade performance. Validated on a single-cohort and three external datasets, VERN showed robust predictive performance and generalizability, providing an open platform (http://plr.20210706.xyz:5000/) to enhance STAS diagnosis efficiency and accuracy.
ESM-NBR: fast and accurate nucleic acid-binding residue prediction via protein language model feature representation and multi-task learning
Dec 01, 2023Abstract:Protein-nucleic acid interactions play a very important role in a variety of biological activities. Accurate identification of nucleic acid-binding residues is a critical step in understanding the interaction mechanisms. Although many computationally based methods have been developed to predict nucleic acid-binding residues, challenges remain. In this study, a fast and accurate sequence-based method, called ESM-NBR, is proposed. In ESM-NBR, we first use the large protein language model ESM2 to extract discriminative biological properties feature representation from protein primary sequences; then, a multi-task deep learning model composed of stacked bidirectional long short-term memory (BiLSTM) and multi-layer perceptron (MLP) networks is employed to explore common and private information of DNA- and RNA-binding residues with ESM2 feature as input. Experimental results on benchmark data sets demonstrate that the prediction performance of ESM2 feature representation comprehensively outperforms evolutionary information-based hidden Markov model (HMM) features. Meanwhile, the ESM-NBR obtains the MCC values for DNA-binding residues prediction of 0.427 and 0.391 on two independent test sets, which are 18.61 and 10.45% higher than those of the second-best methods, respectively. Moreover, by completely discarding the time-cost multiple sequence alignment process, the prediction speed of ESM-NBR far exceeds that of existing methods (5.52s for a protein sequence of length 500, which is about 16 times faster than the second-fastest method). A user-friendly standalone package and the data of ESM-NBR are freely available for academic use at: https://github.com/wwzll123/ESM-NBR.
A New Hybrid Control Architecture to Attenuate Large Horizontal Wind Disturbance for a Small-Scale Unmanned Helicopter
May 03, 2019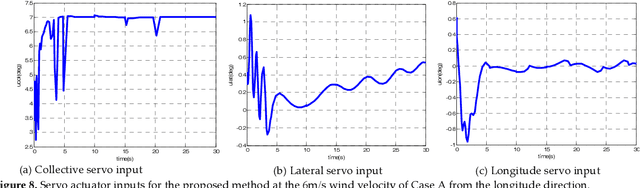
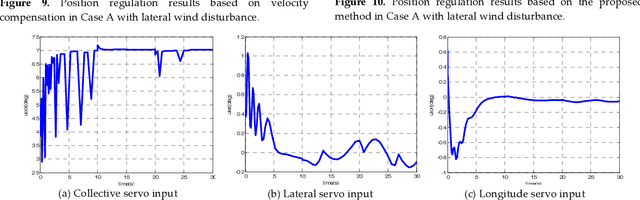
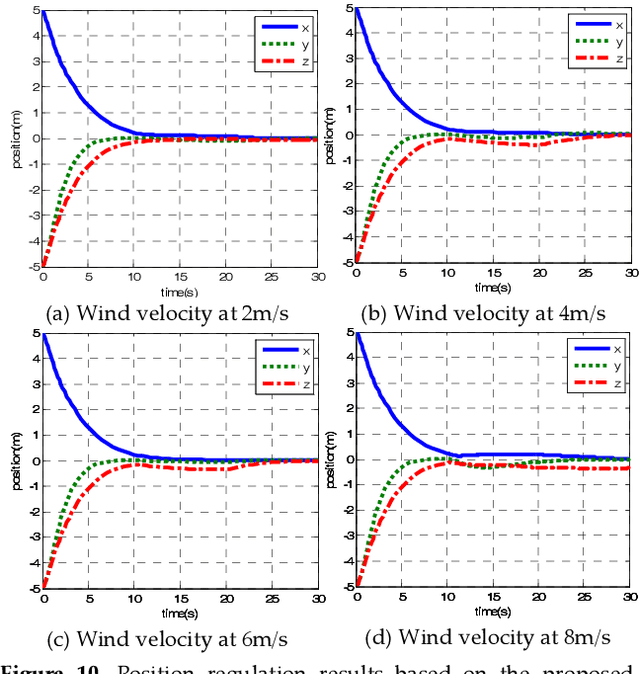
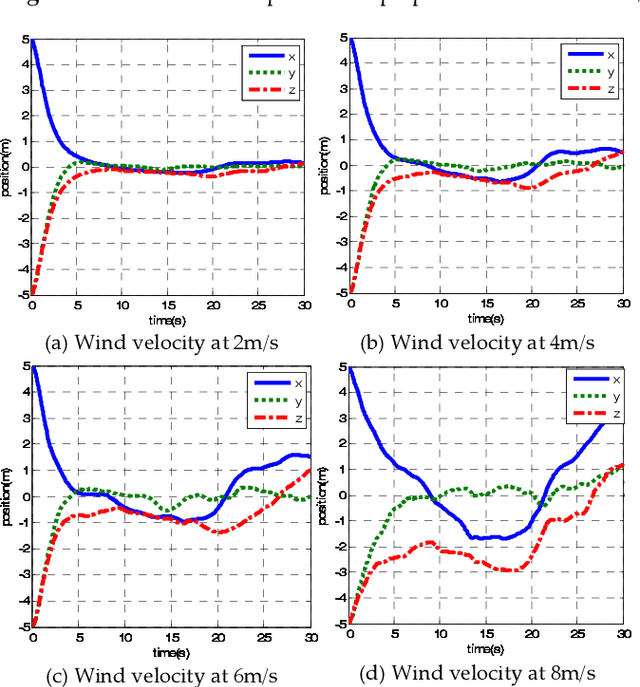
Abstract:This paper presents a novel method to attenuate large horizontal wind disturbance for a small-scale unmanned autonomous helicopter combining wind tunnel-based experimental data and a backstepping algorithm. Large horizontal wind disturbance is harmful to autonomous helicopters, especially to small ones because of their low inertia and the high cross-coupling effects among the multiple inputs. In order to achieve more accurate and faster attenuation of large wind disturbance, a new hybrid control architecture is proposed to take advantage of the direct force/moment compensation based on the wind tunnel experimental data. In this architecture, large horizontal wind disturbance is treated as an additional input to the control system instead of a small perturbation around the equilibrium state. A backstepping algorithm is then designed to guarantee the stable convergence of the hilicopter to the desired position. The proposed technique is finally evaluated in simulation on the platform, HIROBO Eagle, compared with a traditional wind velocity compensation method.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge