Qiannan Zhang
Augmented Risk Prediction for the Onset of Alzheimer's Disease from Electronic Health Records with Large Language Models
May 26, 2024



Abstract:Alzheimer's disease (AD) is the fifth-leading cause of death among Americans aged 65 and older. Screening and early detection of AD and related dementias (ADRD) are critical for timely intervention and for identifying clinical trial participants. The widespread adoption of electronic health records (EHRs) offers an important resource for developing ADRD screening tools such as machine learning based predictive models. Recent advancements in large language models (LLMs) demonstrate their unprecedented capability of encoding knowledge and performing reasoning, which offers them strong potential for enhancing risk prediction. This paper proposes a novel pipeline that augments risk prediction by leveraging the few-shot inference power of LLMs to make predictions on cases where traditional supervised learning methods (SLs) may not excel. Specifically, we develop a collaborative pipeline that combines SLs and LLMs via a confidence-driven decision-making mechanism, leveraging the strengths of SLs in clear-cut cases and LLMs in more complex scenarios. We evaluate this pipeline using a real-world EHR data warehouse from Oregon Health \& Science University (OHSU) Hospital, encompassing EHRs from over 2.5 million patients and more than 20 million patient encounters. Our results show that our proposed approach effectively combines the power of SLs and LLMs, offering significant improvements in predictive performance. This advancement holds promise for revolutionizing ADRD screening and early detection practices, with potential implications for better strategies of patient management and thus improving healthcare.
Joint Analysis of Single-Cell Data across Cohorts with Missing Modalities
May 18, 2024



Abstract:Joint analysis of multi-omic single-cell data across cohorts has significantly enhanced the comprehensive analysis of cellular processes. However, most of the existing approaches for this purpose require access to samples with complete modality availability, which is impractical in many real-world scenarios. In this paper, we propose (Single-Cell Cross-Cohort Cross-Category) integration, a novel framework that learns unified cell representations under domain shift without requiring full-modality reference samples. Our generative approach learns rich cross-modal and cross-domain relationships that enable imputation of these missing modalities. Through experiments on real-world multi-omic datasets, we demonstrate that offers a robust solution to single-cell tasks such as cell type clustering, cell type classification, and feature imputation.
Collaborative Graph Walk for Semi-supervised Multi-Label Node Classification
Nov 17, 2019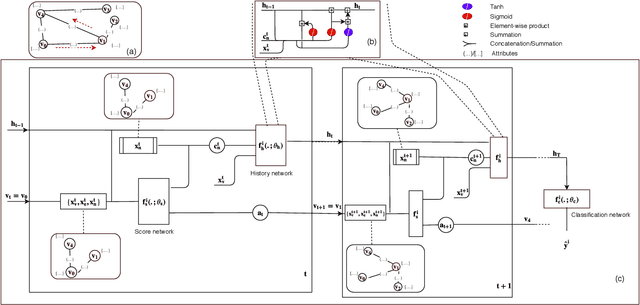
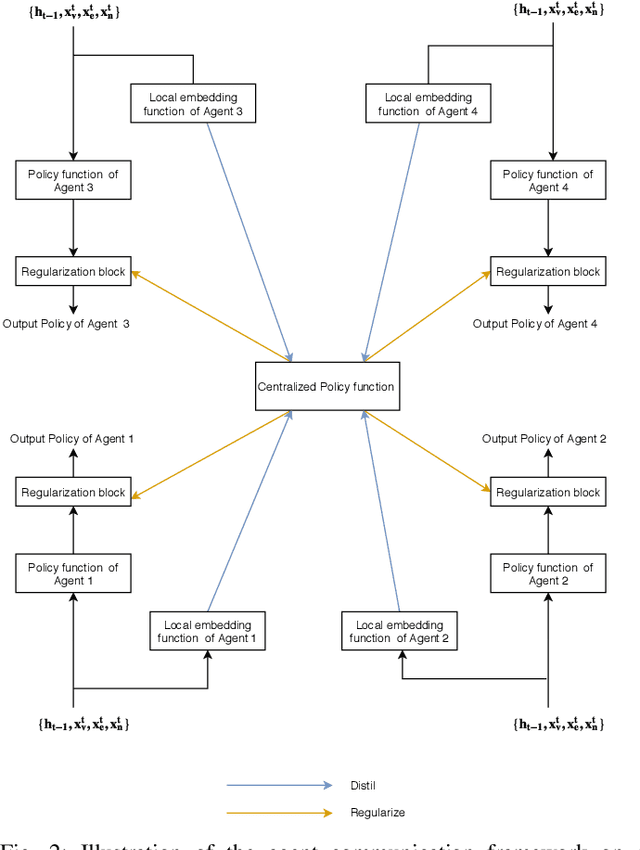
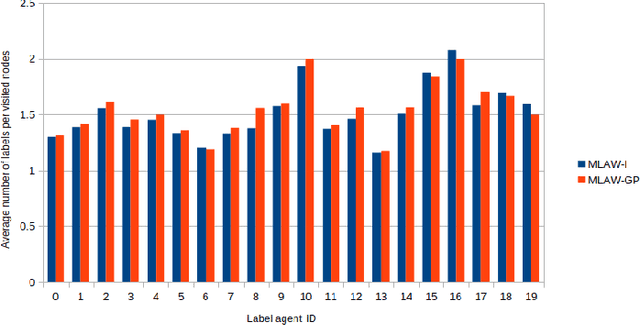
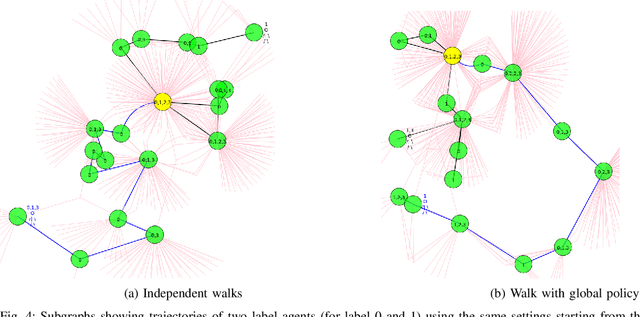
Abstract:In this work, we study semi-supervised multi-label node classification problem in attributed graphs. Classic solutions to multi-label node classification follow two steps, first learn node embedding and then build a node classifier on the learned embedding. To improve the discriminating power of the node embedding, we propose a novel collaborative graph walk, named Multi-Label-Graph-Walk, to finely tune node representations with the available label assignments in attributed graphs via reinforcement learning. The proposed method formulates the multi-label node classification task as simultaneous graph walks conducted by multiple label-specific agents. Furthermore, policies of the label-wise graph walks are learned in a cooperative way to capture first the predictive relation between node labels and structural attributes of graphs; and second, the correlation among the multiple label-specific classification tasks. A comprehensive experimental study demonstrates that the proposed method can achieve significantly better multi-label classification performance than the state-of-the-art approaches and conduct more efficient graph exploration.
Recurrent Attention Walk for Semi-supervised Classification
Oct 22, 2019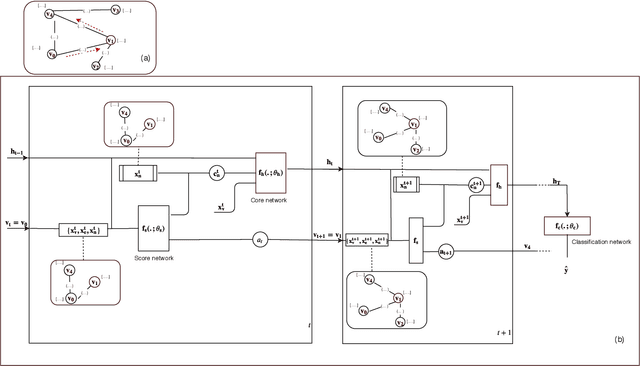
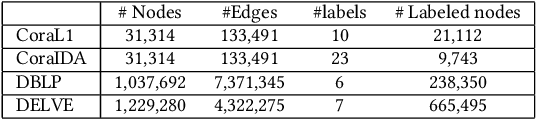
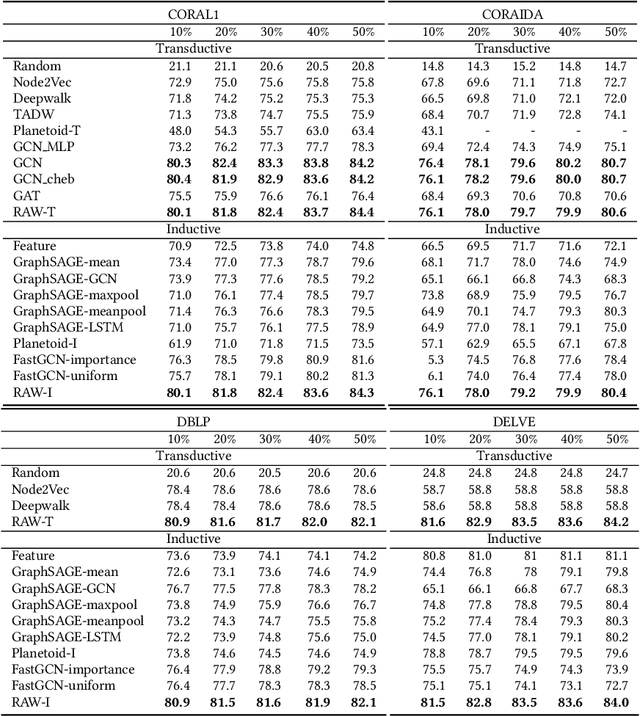
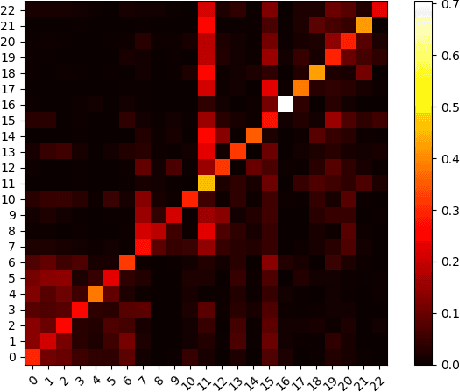
Abstract:In this paper, we study the graph-based semi-supervised learning for classifying nodes in attributed networks, where the nodes and edges possess content information. Recent approaches like graph convolution networks and attention mechanisms have been proposed to ensemble the first-order neighbors and incorporate the relevant neighbors. However, it is costly (especially in memory) to consider all neighbors without a prior differentiation. We propose to explore the neighborhood in a reinforcement learning setting and find a walk path well-tuned for classifying the unlabelled target nodes. We let an agent (of node classification task) walk over the graph and decide where to direct to maximize classification accuracy. We define the graph walk as a partially observable Markov decision process (POMDP). The proposed method is flexible for working in both transductive and inductive setting. Extensive experiments on four datasets demonstrate that our proposed method outperforms several state-of-the-art methods. Several case studies also illustrate the meaningful movement trajectory made by the agent.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge