Han Yufei
Permutation-Invariant Subgraph Discovery
Apr 02, 2021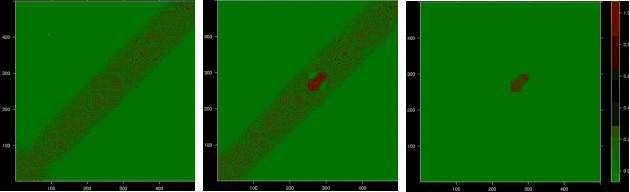
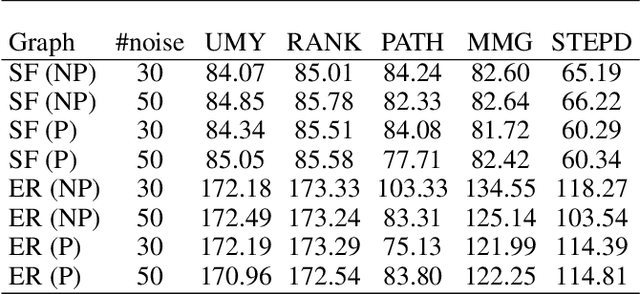
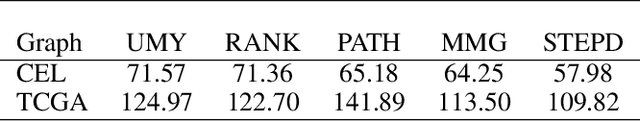
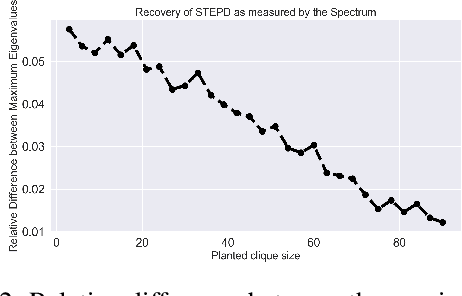
Abstract:We introduce Permutation and Structured Perturbation Inference (PSPI), a new problem formulation that abstracts many graph matching tasks that arise in systems biology. PSPI can be viewed as a robust formulation of the permutation inference or graph matching, where the objective is to find a permutation between two graphs under the assumption that a set of edges may have undergone a perturbation due to an underlying cause. For example, suppose there are two gene regulatory networks X and Y from a diseased and normal tissue respectively. Then, the PSPI problem can be used to detect if there has been a structural change between the two networks which can serve as a signature of the disease. Besides the new problem formulation, we propose an ADMM algorithm (STEPD) to solve a relaxed version of the PSPI problem. An extensive case study on comparative gene regulatory networks (GRNs) is used to demonstrate that STEPD is able to accurately infer structured perturbations and thus provides a tool for computational biologists to identify novel prognostic signatures. A spectral analysis confirms that STEPD can recover small clique-like perturbations making it a useful tool for detecting permutation-invariant changes in graphs.
Collaborative Graph Walk for Semi-supervised Multi-Label Node Classification
Nov 17, 2019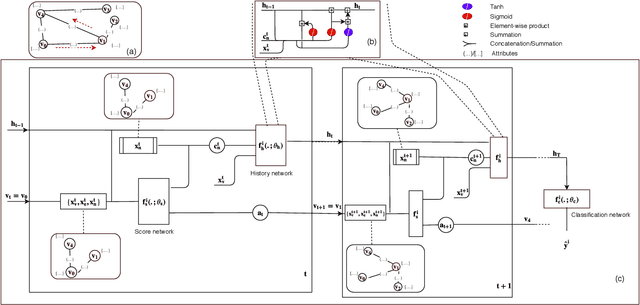
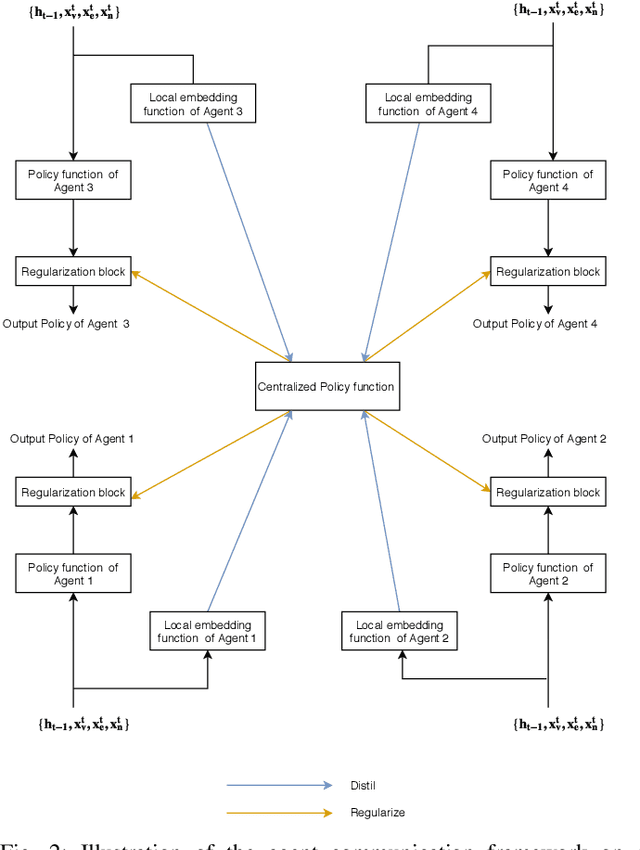
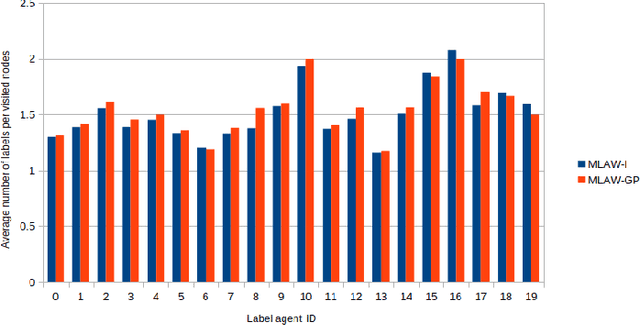
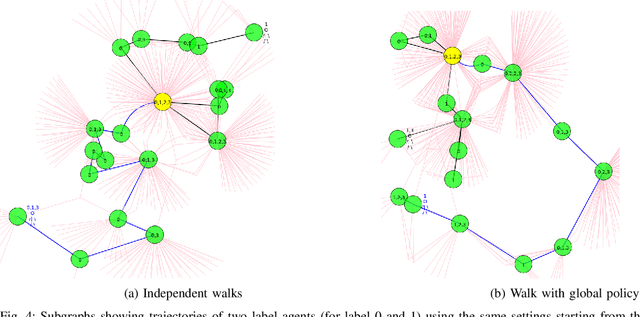
Abstract:In this work, we study semi-supervised multi-label node classification problem in attributed graphs. Classic solutions to multi-label node classification follow two steps, first learn node embedding and then build a node classifier on the learned embedding. To improve the discriminating power of the node embedding, we propose a novel collaborative graph walk, named Multi-Label-Graph-Walk, to finely tune node representations with the available label assignments in attributed graphs via reinforcement learning. The proposed method formulates the multi-label node classification task as simultaneous graph walks conducted by multiple label-specific agents. Furthermore, policies of the label-wise graph walks are learned in a cooperative way to capture first the predictive relation between node labels and structural attributes of graphs; and second, the correlation among the multiple label-specific classification tasks. A comprehensive experimental study demonstrates that the proposed method can achieve significantly better multi-label classification performance than the state-of-the-art approaches and conduct more efficient graph exploration.
Recurrent Attention Walk for Semi-supervised Classification
Oct 22, 2019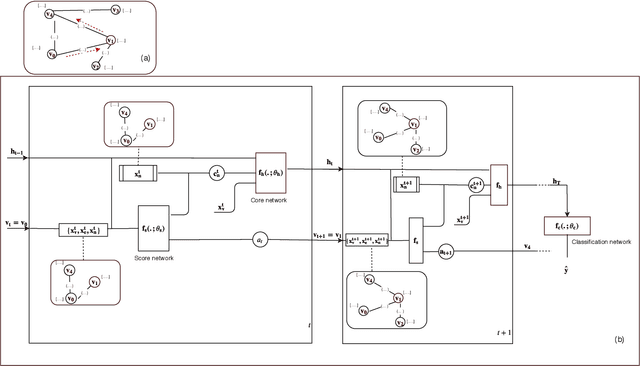
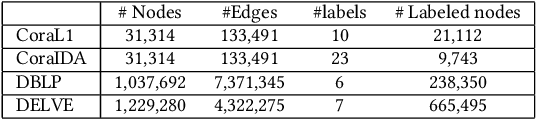
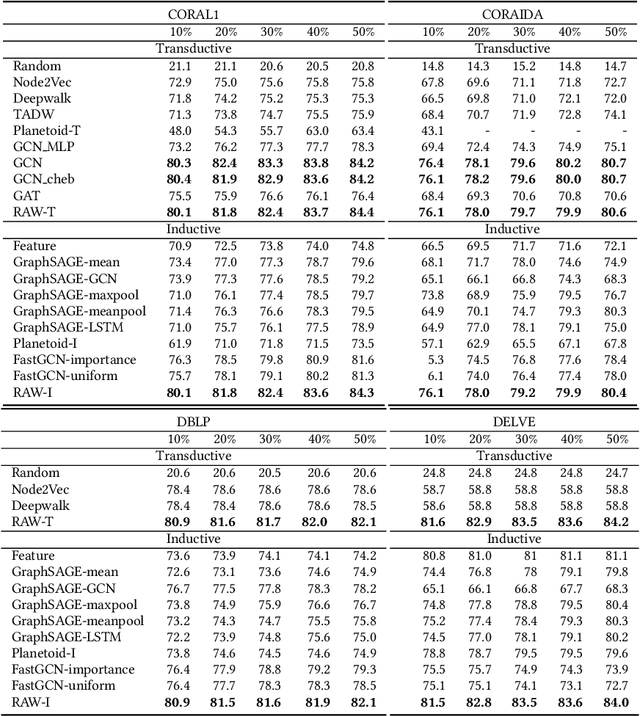
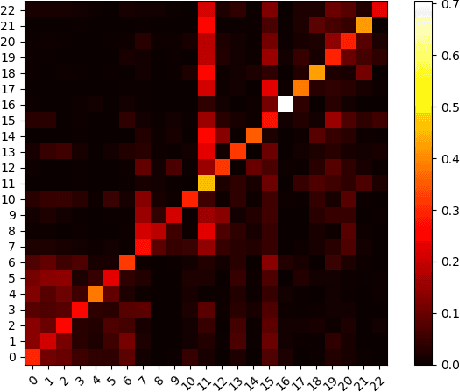
Abstract:In this paper, we study the graph-based semi-supervised learning for classifying nodes in attributed networks, where the nodes and edges possess content information. Recent approaches like graph convolution networks and attention mechanisms have been proposed to ensemble the first-order neighbors and incorporate the relevant neighbors. However, it is costly (especially in memory) to consider all neighbors without a prior differentiation. We propose to explore the neighborhood in a reinforcement learning setting and find a walk path well-tuned for classifying the unlabelled target nodes. We let an agent (of node classification task) walk over the graph and decide where to direct to maximize classification accuracy. We define the graph walk as a partially observable Markov decision process (POMDP). The proposed method is flexible for working in both transductive and inductive setting. Extensive experiments on four datasets demonstrate that our proposed method outperforms several state-of-the-art methods. Several case studies also illustrate the meaningful movement trajectory made by the agent.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge