Uchenna Akujuobi
MEG: Medical Knowledge-Augmented Large Language Models for Question Answering
Nov 07, 2024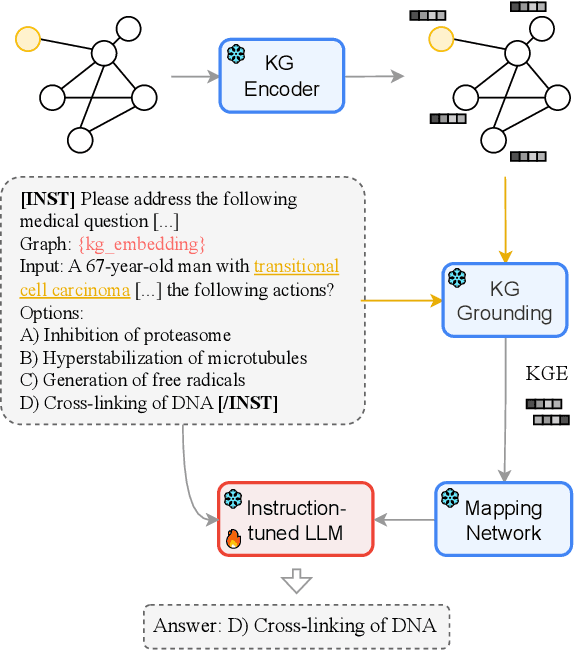

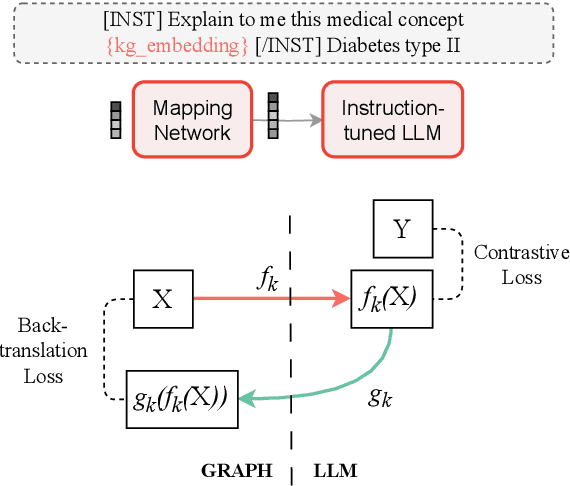

Abstract:Question answering is a natural language understanding task that involves reasoning over both explicit context and unstated, relevant domain knowledge. Large language models (LLMs), which underpin most contemporary question answering systems, struggle to induce how concepts relate in specialized domains such as medicine. Existing medical LLMs are also costly to train. In this work, we present MEG, a parameter-efficient approach for medical knowledge-augmented LLMs. MEG uses a lightweight mapping network to integrate graph embeddings into the LLM, enabling it to leverage external knowledge in a cost-effective way. We evaluate our method on four popular medical multiple-choice datasets and show that LLMs greatly benefit from the factual grounding provided by knowledge graph embeddings. MEG attains an average of +10.2% accuracy over the Mistral-Instruct baseline, and +6.7% over specialized models like BioMistral. We also show results based on Llama-3. Finally, we show that MEG's performance remains robust to the choice of graph encoder.
It is Simple Sometimes: A Study On Improving Aspect-Based Sentiment Analysis Performance
May 31, 2024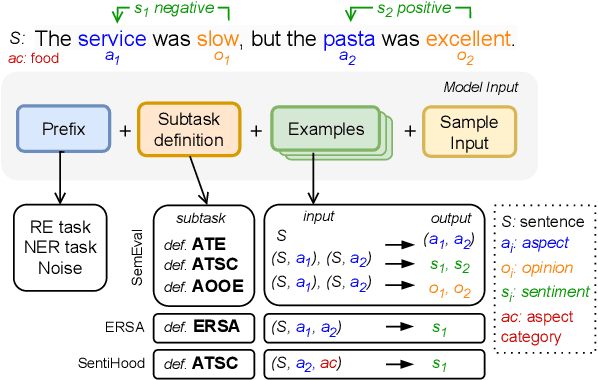
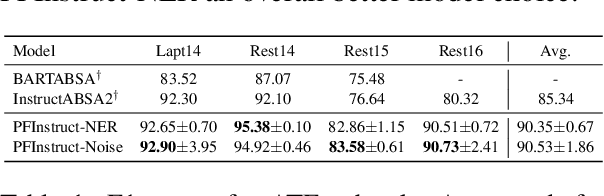
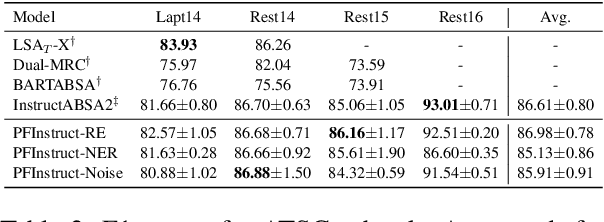
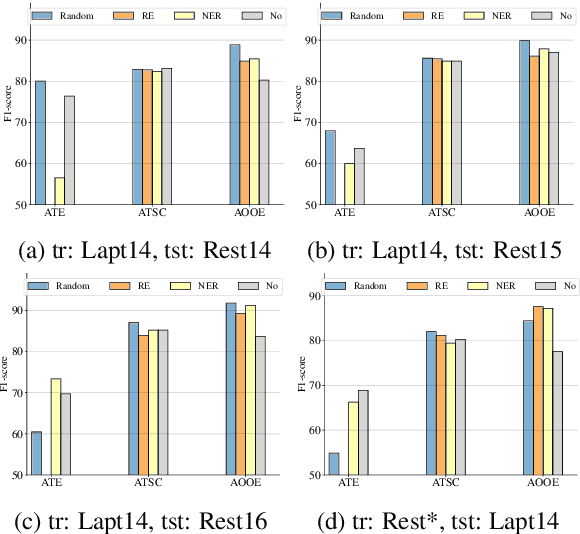
Abstract:Aspect-Based Sentiment Analysis (ABSA) involves extracting opinions from textual data about specific entities and their corresponding aspects through various complementary subtasks. Several prior research has focused on developing ad hoc designs of varying complexities for these subtasks. In this paper, we present a generative framework extensible to any ABSA subtask. We build upon the instruction tuned model proposed by Scaria et al. (2023), who present an instruction-based model with task descriptions followed by in-context examples on ABSA subtasks. We propose PFInstruct, an extension to this instruction learning paradigm by appending an NLP-related task prefix to the task description. This simple approach leads to improved performance across all tested SemEval subtasks, surpassing previous state-of-the-art (SOTA) on the ATE subtask (Rest14) by +3.28 F1-score, and on the AOOE subtask by an average of +5.43 F1-score across SemEval datasets. Furthermore, we explore the impact of the prefix-enhanced prompt quality on the ABSA subtasks and find that even a noisy prefix enhances model performance compared to the baseline. Our method also achieves competitive results on a biomedical domain dataset (ERSA).
CERM: Context-aware Literature-based Discovery via Sentiment Analysis
Jan 27, 2024Abstract:Driven by the abundance of biomedical publications, we introduce a sentiment analysis task to understand food-health relationship. Prior attempts to incorporate health into recipe recommendation and analysis systems have primarily focused on ingredient nutritional components or utilized basic computational models trained on curated labeled data. Enhanced models that capture the inherent relationship between food ingredients and biomedical concepts can be more beneficial for food-related research, given the wealth of information in biomedical texts. Considering the costly data labeling process, these models should effectively utilize both labeled and unlabeled data. This paper introduces Entity Relationship Sentiment Analysis (ERSA), a new task that captures the sentiment of a text based on an entity pair. ERSA extends the widely studied Aspect Based Sentiment Analysis (ABSA) task. Specifically, our study concentrates on the ERSA task applied to biomedical texts, focusing on (entity-entity) pairs of biomedical and food concepts. ERSA poses a significant challenge compared to traditional sentiment analysis tasks, as sentence sentiment may not align with entity relationship sentiment. Additionally, we propose CERM, a semi-supervised architecture that combines different word embeddings to enhance the encoding of the ERSA task. Experimental results showcase the model's efficiency across diverse learning scenarios.
Temporal Positive-unlabeled Learning for Biomedical Hypothesis Generation via Risk Estimation
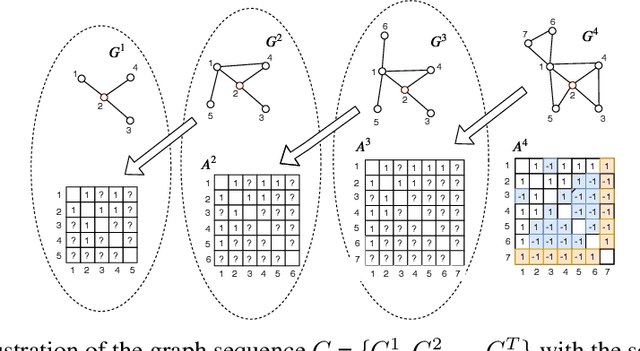
Oct 05, 2020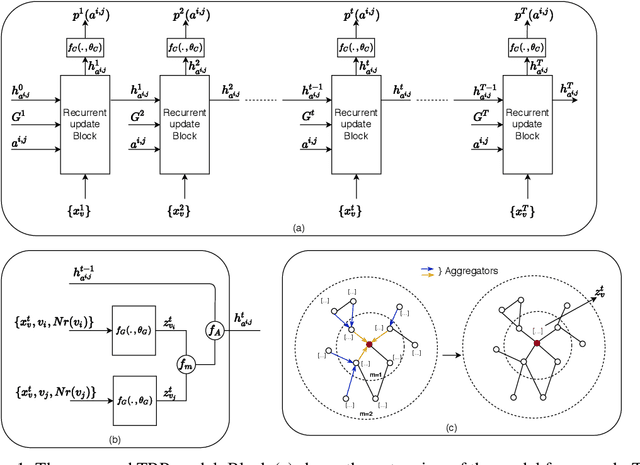

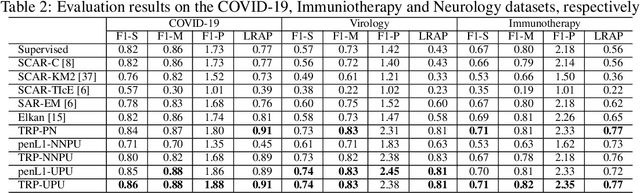
Abstract:Understanding the relationships between biomedical terms like viruses, drugs, and symptoms is essential in the fight against diseases. Many attempts have been made to introduce the use of machine learning to the scientific process of hypothesis generation(HG), which refers to the discovery of meaningful implicit connections between biomedical terms. However, most existing methods fail to truly capture the temporal dynamics of scientific term relations and also assume unobserved connections to be irrelevant (i.e., in a positive-negative (PN) learning setting). To break these limits, we formulate this HG problem as future connectivity prediction task on a dynamic attributed graph via positive-unlabeled (PU) learning. Then, the key is to capture the temporal evolution of node pair (term pair) relations from just the positive and unlabeled data. We propose a variational inference model to estimate the positive prior, and incorporate it in the learning of node pair embeddings, which are then used for link prediction. Experiment results on real-world biomedical term relationship datasets and case study analyses on a COVID-19 dataset validate the effectiveness of the proposed model.
Collaborative Graph Walk for Semi-supervised Multi-Label Node Classification
Nov 17, 2019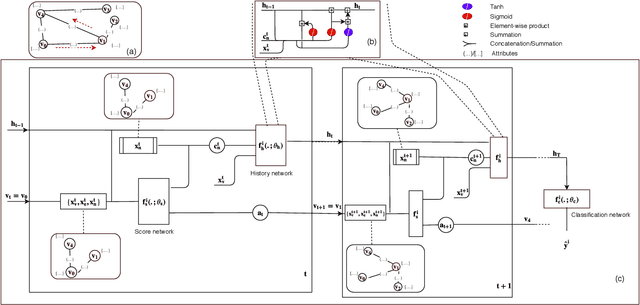
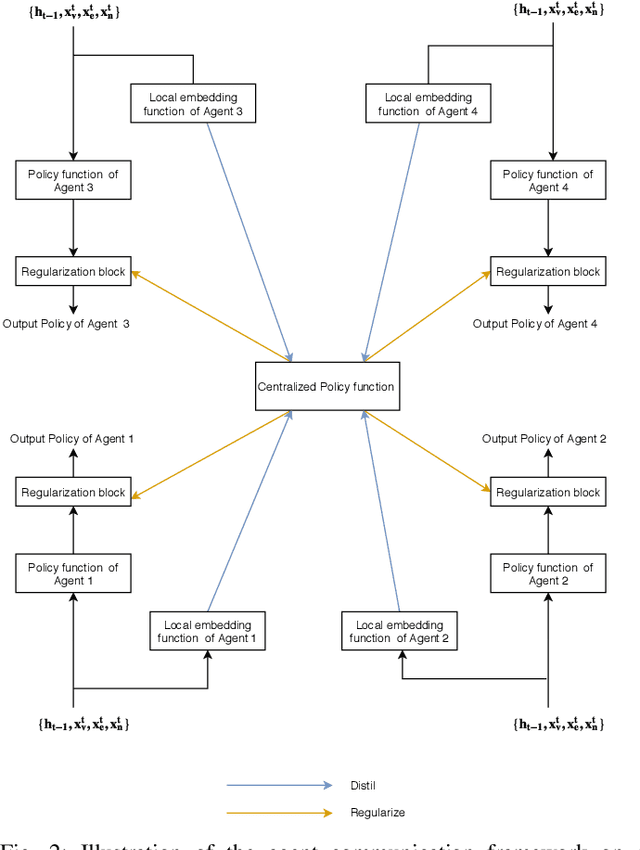
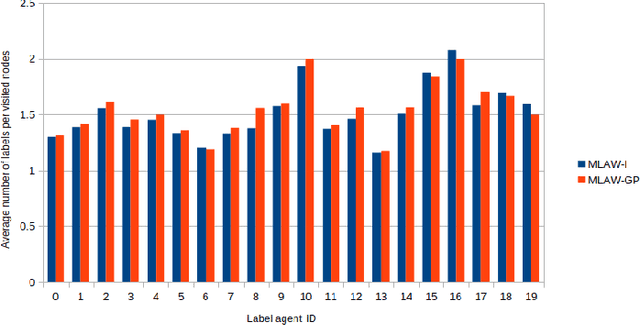
Abstract:In this work, we study semi-supervised multi-label node classification problem in attributed graphs. Classic solutions to multi-label node classification follow two steps, first learn node embedding and then build a node classifier on the learned embedding. To improve the discriminating power of the node embedding, we propose a novel collaborative graph walk, named Multi-Label-Graph-Walk, to finely tune node representations with the available label assignments in attributed graphs via reinforcement learning. The proposed method formulates the multi-label node classification task as simultaneous graph walks conducted by multiple label-specific agents. Furthermore, policies of the label-wise graph walks are learned in a cooperative way to capture first the predictive relation between node labels and structural attributes of graphs; and second, the correlation among the multiple label-specific classification tasks. A comprehensive experimental study demonstrates that the proposed method can achieve significantly better multi-label classification performance than the state-of-the-art approaches and conduct more efficient graph exploration.
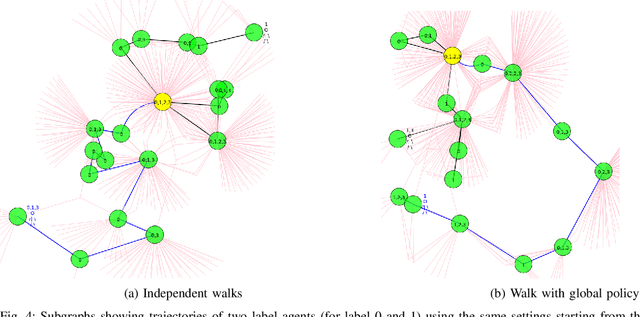
Recurrent Attention Walk for Semi-supervised Classification
Oct 22, 2019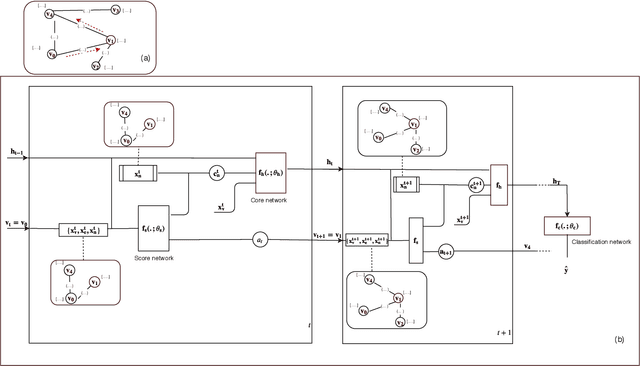
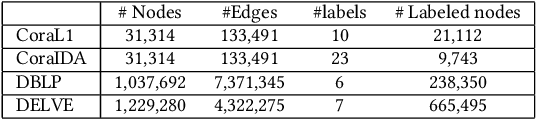
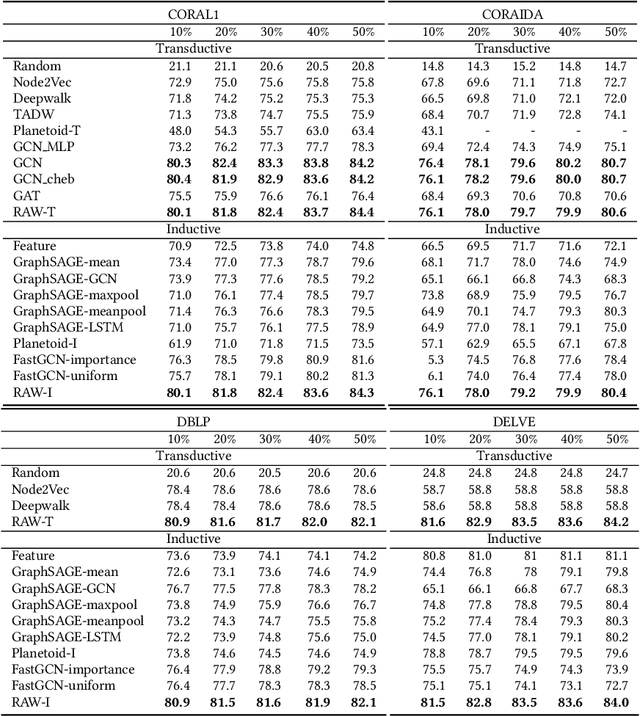
Abstract:In this paper, we study the graph-based semi-supervised learning for classifying nodes in attributed networks, where the nodes and edges possess content information. Recent approaches like graph convolution networks and attention mechanisms have been proposed to ensemble the first-order neighbors and incorporate the relevant neighbors. However, it is costly (especially in memory) to consider all neighbors without a prior differentiation. We propose to explore the neighborhood in a reinforcement learning setting and find a walk path well-tuned for classifying the unlabelled target nodes. We let an agent (of node classification task) walk over the graph and decide where to direct to maximize classification accuracy. We define the graph walk as a partially observable Markov decision process (POMDP). The proposed method is flexible for working in both transductive and inductive setting. Extensive experiments on four datasets demonstrate that our proposed method outperforms several state-of-the-art methods. Several case studies also illustrate the meaningful movement trajectory made by the agent.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge