Pingjun Chen
CellSymphony: Deciphering the molecular and phenotypic orchestration of cells with single-cell pathomics
Aug 13, 2025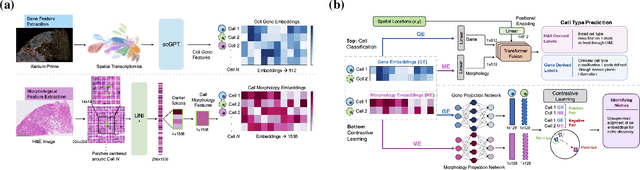
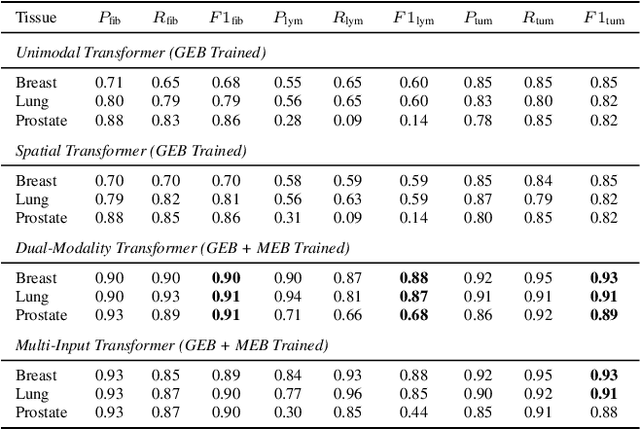
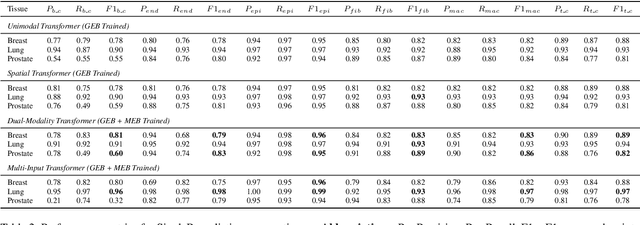
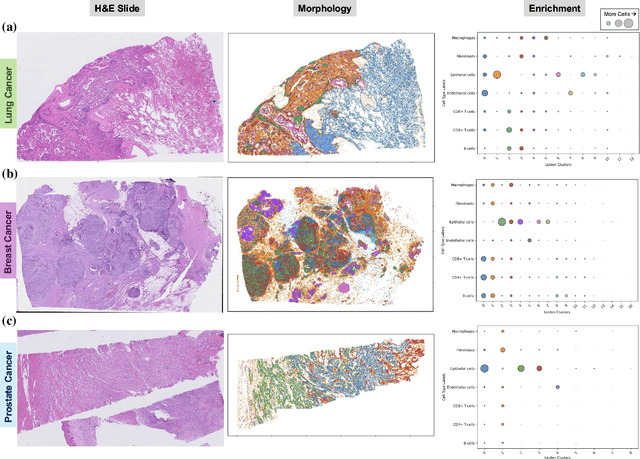
Abstract:Xenium, a new spatial transcriptomics platform, enables subcellular-resolution profiling of complex tumor tissues. Despite the rich morphological information in histology images, extracting robust cell-level features and integrating them with spatial transcriptomics data remains a critical challenge. We introduce CellSymphony, a flexible multimodal framework that leverages foundation model-derived embeddings from both Xenium transcriptomic profiles and histology images at true single-cell resolution. By learning joint representations that fuse spatial gene expression with morphological context, CellSymphony achieves accurate cell type annotation and uncovers distinct microenvironmental niches across three cancer types. This work highlights the potential of foundation models and multimodal fusion for deciphering the physiological and phenotypic orchestration of cells within complex tissue ecosystems.
Hierarchical Phenotyping and Graph Modeling of Spatial Architecture in Lymphoid Neoplasms
Jun 30, 2021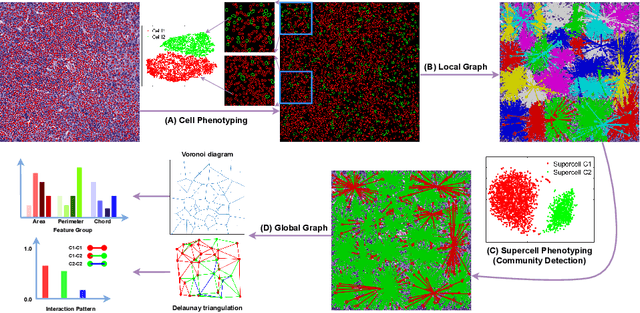
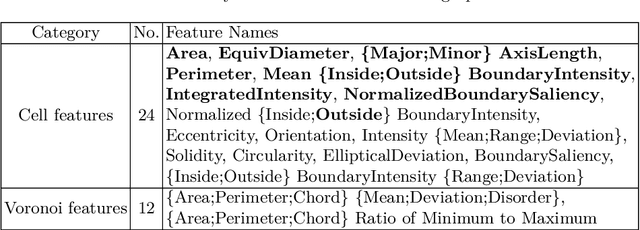
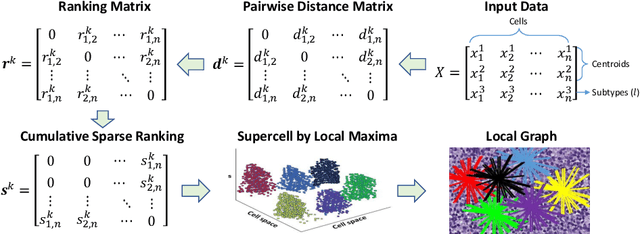
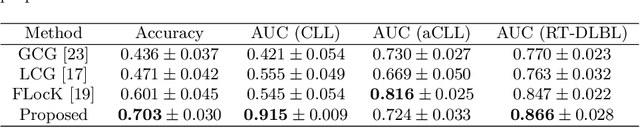
Abstract:The cells and their spatial patterns in the tumor microenvironment (TME) play a key role in tumor evolution, and yet remains an understudied topic in computational pathology. This study, to the best of our knowledge, is among the first to hybrid local and global graph methods to profile orchestration and interaction of cellular components. To address the challenge in hematolymphoid cancers where the cell classes in TME are unclear, we first implemented cell level unsupervised learning and identified two new cell subtypes. Local cell graphs or supercells were built for each image by considering the individual cell's geospatial location and classes. Then, we applied supercell level clustering and identified two new cell communities. In the end, we built global graphs to abstract spatial interaction patterns and extract features for disease diagnosis. We evaluate the proposed algorithm on H\&E slides of 60 hematolymphoid neoplasm patients and further compared it with three cell level graph-based algorithms, including the global cell graph, cluster cell graph, and FLocK. The proposed algorithm achieves a mean diagnosis accuracy of 0.703 with the repeated 5-fold cross-validation scheme. In conclusion, our algorithm shows superior performance over the existing methods and can be potentially applied to other cancer types.
Anatomy-Aware Cardiac Motion Estimation
Aug 17, 2020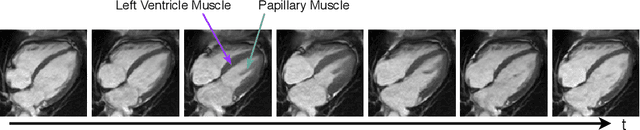
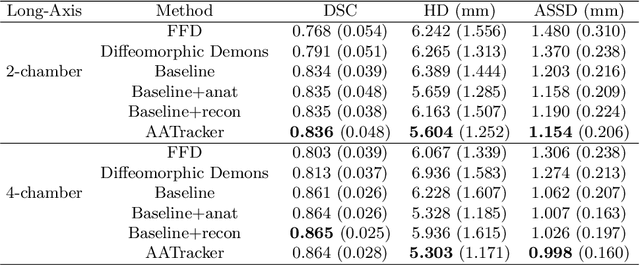
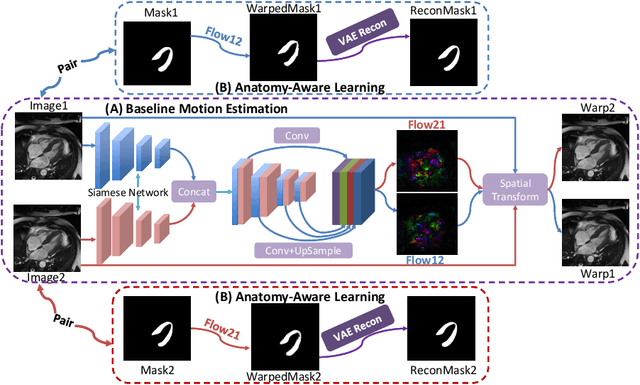
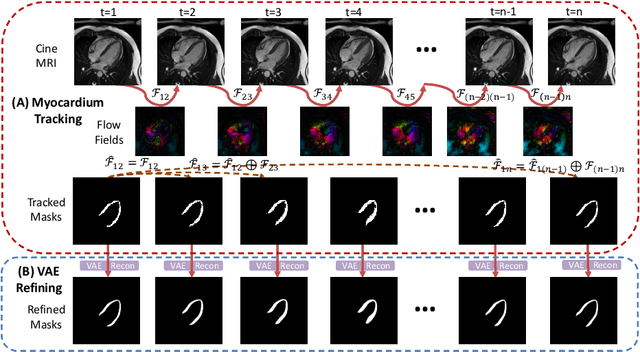
Abstract:Cardiac motion estimation is critical to the assessment of cardiac function. Myocardium feature tracking (FT) can directly estimate cardiac motion from cine MRI, which requires no special scanning procedure. However, current deep learning-based FT methods may result in unrealistic myocardium shapes since the learning is solely guided by image intensities without considering anatomy. On the other hand, motion estimation through learning is challenging because ground-truth motion fields are almost impossible to obtain. In this study, we propose a novel Anatomy-Aware Tracker (AATracker) for cardiac motion estimation that preserves anatomy by weak supervision. A convolutional variational autoencoder (VAE) is trained to encapsulate realistic myocardium shapes. A baseline dense motion tracker is trained to approximate the motion fields and then refined to estimate anatomy-aware motion fields under the weak supervision from the VAE. We evaluate the proposed method on long-axis cardiac cine MRI, which has more complex myocardium appearances and motions than short-axis. Compared with other methods, AATracker significantly improves the tracking performance and provides visually more realistic tracking results, demonstrating the effectiveness of the proposed weakly-supervision scheme in cardiac motion estimation.
Iterative Attention Mining for Weakly Supervised Thoracic Disease Pattern Localization in Chest X-Rays
Jul 03, 2018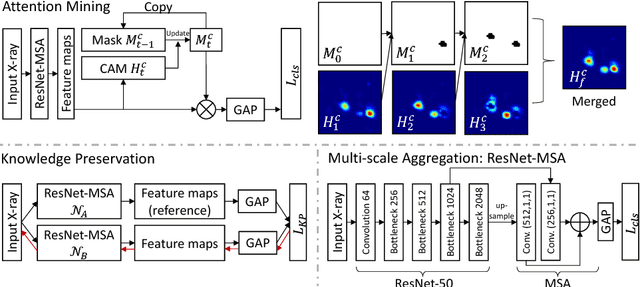
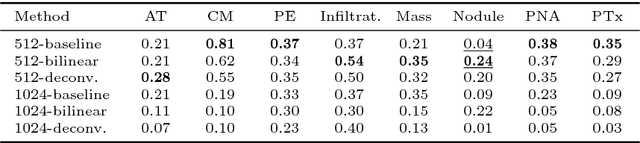
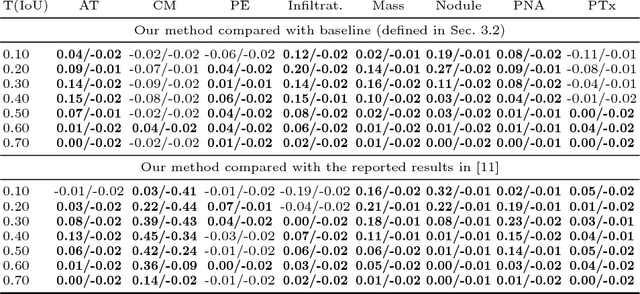
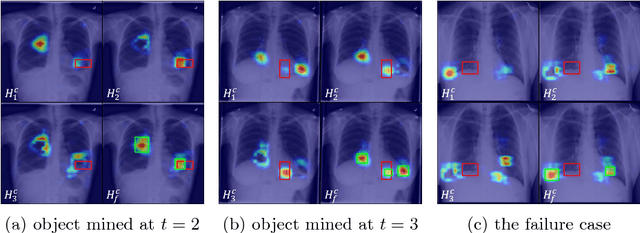
Abstract:Given image labels as the only supervisory signal, we focus on harvesting, or mining, thoracic disease localizations from chest X-ray images. Harvesting such localizations from existing datasets allows for the creation of improved data sources for computer-aided diagnosis and retrospective analyses. We train a convolutional neural network (CNN) for image classification and propose an attention mining (AM) strategy to improve the model's sensitivity or saliency to disease patterns. The intuition of AM is that once the most salient disease area is blocked or hidden from the CNN model, it will pay attention to alternative image regions, while still attempting to make correct predictions. However, the model requires to be properly constrained during AM, otherwise, it may overfit to uncorrelated image parts and forget the valuable knowledge that it has learned from the original image classification task. To alleviate such side effects, we then design a knowledge preservation (KP) loss, which minimizes the discrepancy between responses for X-ray images from the original and the updated networks. Furthermore, we modify the CNN model to include multi-scale aggregation (MSA), improving its localization ability on small-scale disease findings, e.g., lung nodules. We experimentally validate our method on the publicly-available ChestX-ray14 dataset, outperforming a class activation map (CAM)-based approach, and demonstrating the value of our novel framework for mining disease locations.
TandemNet: Distilling Knowledge from Medical Images Using Diagnostic Reports as Optional Semantic References
Aug 10, 2017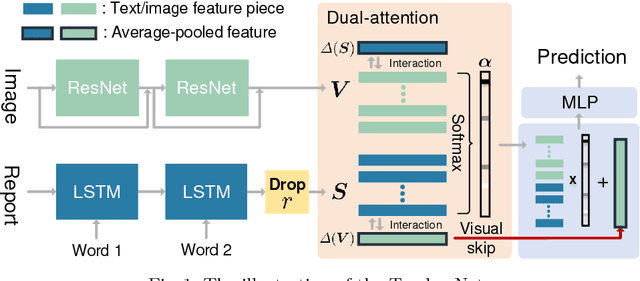
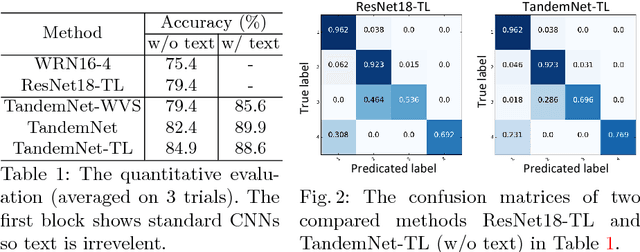
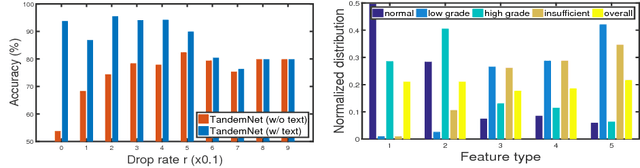

Abstract:In this paper, we introduce the semantic knowledge of medical images from their diagnostic reports to provide an inspirational network training and an interpretable prediction mechanism with our proposed novel multimodal neural network, namely TandemNet. Inside TandemNet, a language model is used to represent report text, which cooperates with the image model in a tandem scheme. We propose a novel dual-attention model that facilitates high-level interactions between visual and semantic information and effectively distills useful features for prediction. In the testing stage, TandemNet can make accurate image prediction with an optional report text input. It also interprets its prediction by producing attention on the image and text informative feature pieces, and further generating diagnostic report paragraphs. Based on a pathological bladder cancer images and their diagnostic reports (BCIDR) dataset, sufficient experiments demonstrate that our method effectively learns and integrates knowledge from multimodalities and obtains significantly improved performance than comparing baselines.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge