Muhammad Aminu
Hierarchical Phenotyping and Graph Modeling of Spatial Architecture in Lymphoid Neoplasms
Jun 30, 2021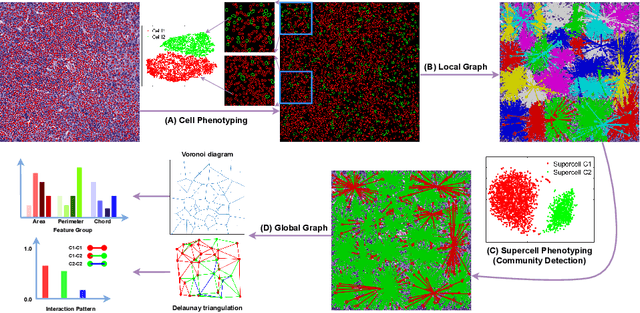
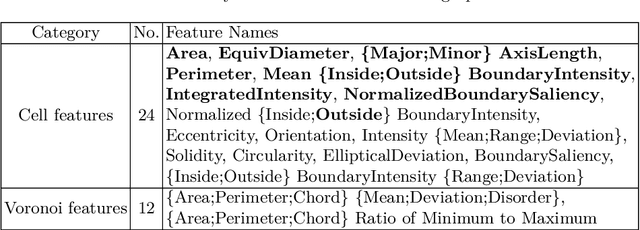
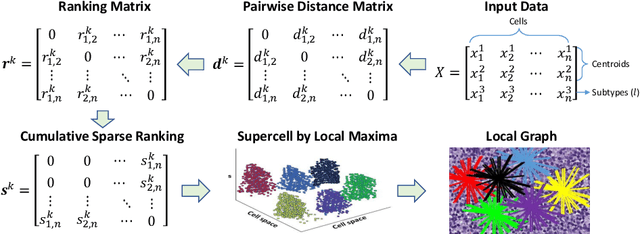
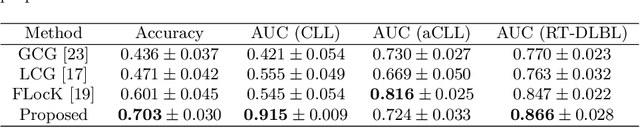
Abstract:The cells and their spatial patterns in the tumor microenvironment (TME) play a key role in tumor evolution, and yet remains an understudied topic in computational pathology. This study, to the best of our knowledge, is among the first to hybrid local and global graph methods to profile orchestration and interaction of cellular components. To address the challenge in hematolymphoid cancers where the cell classes in TME are unclear, we first implemented cell level unsupervised learning and identified two new cell subtypes. Local cell graphs or supercells were built for each image by considering the individual cell's geospatial location and classes. Then, we applied supercell level clustering and identified two new cell communities. In the end, we built global graphs to abstract spatial interaction patterns and extract features for disease diagnosis. We evaluate the proposed algorithm on H\&E slides of 60 hematolymphoid neoplasm patients and further compared it with three cell level graph-based algorithms, including the global cell graph, cluster cell graph, and FLocK. The proposed algorithm achieves a mean diagnosis accuracy of 0.703 with the repeated 5-fold cross-validation scheme. In conclusion, our algorithm shows superior performance over the existing methods and can be potentially applied to other cancer types.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge