Philippe Cattin
GAMER-MRIL identifies Disability-Related Brain Changes in Multiple Sclerosis
Aug 15, 2023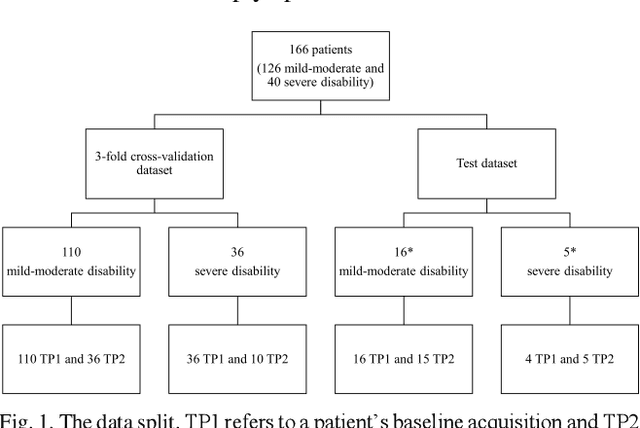
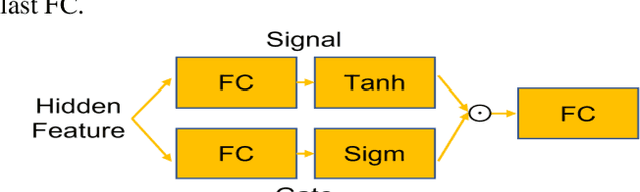
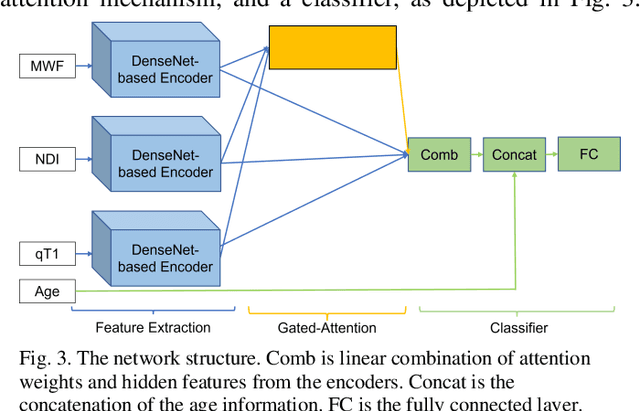
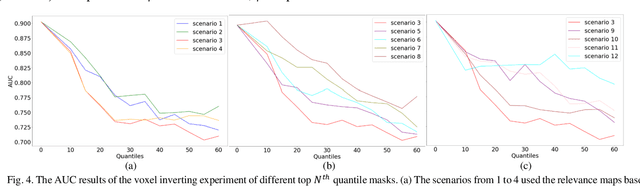
Abstract:Objective: Identifying disability-related brain changes is important for multiple sclerosis (MS) patients. Currently, there is no clear understanding about which pathological features drive disability in single MS patients. In this work, we propose a novel comprehensive approach, GAMER-MRIL, leveraging whole-brain quantitative MRI (qMRI), convolutional neural network (CNN), and an interpretability method from classifying MS patients with severe disability to investigating relevant pathological brain changes. Methods: One-hundred-sixty-six MS patients underwent 3T MRI acquisitions. qMRI informative of microstructural brain properties was reconstructed, including quantitative T1 (qT1), myelin water fraction (MWF), and neurite density index (NDI). To fully utilize the qMRI, GAMER-MRIL extended a gated-attention-based CNN (GAMER-MRI), which was developed to select patch-based qMRI important for a given task/question, to the whole-brain image. To find out disability-related brain regions, GAMER-MRIL modified a structure-aware interpretability method, Layer-wise Relevance Propagation (LRP), to incorporate qMRI. Results: The test performance was AUC=0.885. qT1 was the most sensitive measure related to disability, followed by NDI. The proposed LRP approach obtained more specifically relevant regions than other interpretability methods, including the saliency map, the integrated gradients, and the original LRP. The relevant regions included the corticospinal tract, where average qT1 and NDI significantly correlated with patients' disability scores ($\rho$=-0.37 and 0.44). Conclusion: These results demonstrated that GAMER-MRIL can classify patients with severe disability using qMRI and subsequently identify brain regions potentially important to the integrity of the mobile function. Significance: GAMER-MRIL holds promise for developing biomarkers and increasing clinicians' trust in NN.
Ensemble uncertainty as a criterion for dataset expansion in distinct bone segmentation from upper-body CT images
Aug 19, 2022
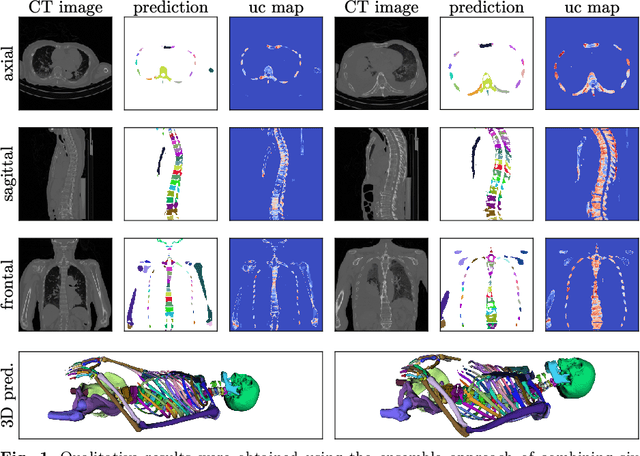
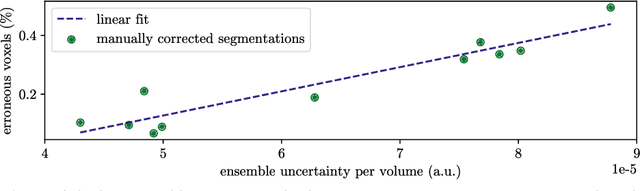
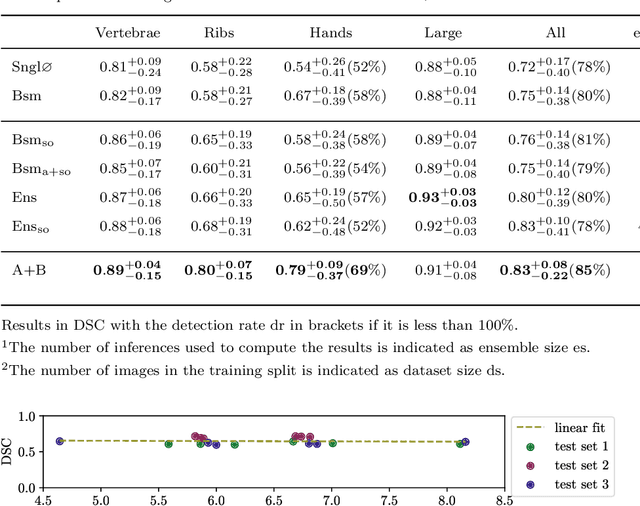
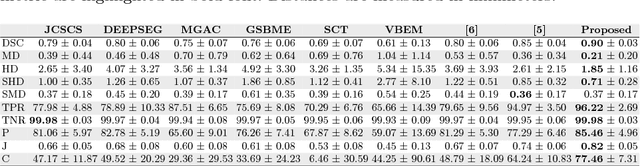
Abstract:Purpose: The localisation and segmentation of individual bones is an important preprocessing step in many planning and navigation applications. It is, however, a time-consuming and repetitive task if done manually. This is true not only for clinical practice but also for the acquisition of training data. We therefore not only present an end-to-end learnt algorithm that is capable of segmenting 125 distinct bones in an upper-body CT, but also provide an ensemble-based uncertainty measure that helps to single out scans to enlarge the training dataset with. Methods We create fully automated end-to-end learnt segmentations using a neural network architecture inspired by the 3D-Unet and fully supervised training. The results are improved using ensembles and inference-time augmentation. We examine the relationship of ensemble-uncertainty to an unlabelled scan's prospective usefulness as part of the training dataset. Results: Our methods are evaluated on an in-house dataset of 16 upper-body CT scans with a resolution of \SI{2}{\milli\meter} per dimension. Taking into account all 125 bones in our label set, our most successful ensemble achieves a median dice score coefficient of 0.83. We find a lack of correlation between a scan's ensemble uncertainty and its prospective influence on the accuracies achieved within an enlarged training set. At the same time, we show that the ensemble uncertainty correlates to the number of voxels that need manual correction after an initial automated segmentation, thus minimising the time required to finalise a new ground truth segmentation. Conclusion: In combination, scans with low ensemble uncertainty need less annotator time while yielding similar future DSC improvements. They are thus ideal candidates to enlarge a training set for upper-body distinct bone segmentation from CT scans. }
Spinal Cord Gray Matter-White Matter Segmentation on Magnetic Resonance AMIRA Images with MD-GRU
Aug 07, 2018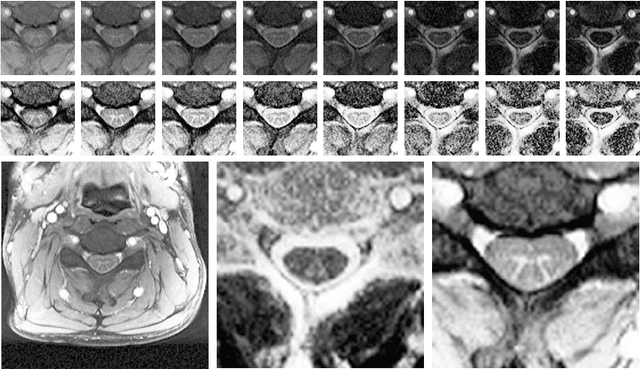
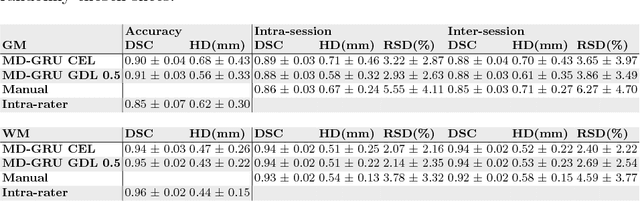
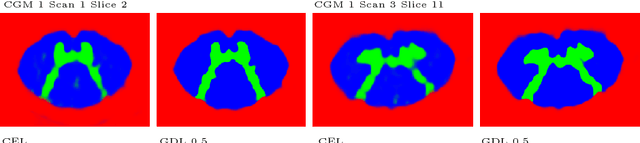
Abstract:The small butterfly shaped structure of spinal cord (SC) gray matter (GM) is challenging to image and to delinate from its surrounding white matter (WM). Segmenting GM is up to a point a trade-off between accuracy and precision. We propose a new pipeline for GM-WM magnetic resonance (MR) image acquisition and segmentation. We report superior results as compared to the ones recently reported in the SC GM segmentation challenge and show even better results using the averaged magnetization inversion recovery acquisitions (AMIRA) sequence. Scan-rescan experiments with the AMIRA sequence show high reproducibility in terms of Dice coefficient, Hausdorff distance and relative standard deviation. We use a recurrent neural network (RNN) with multi-dimensional gated recurrent units (MD-GRU) to train segmentation models on the AMIRA dataset of 855 slices. We added a generalized dice loss to the cross entropy loss that MD-GRU uses and were able to improve the results.
Pathology Segmentation using Distributional Differences to Images of Healthy Origin
May 25, 2018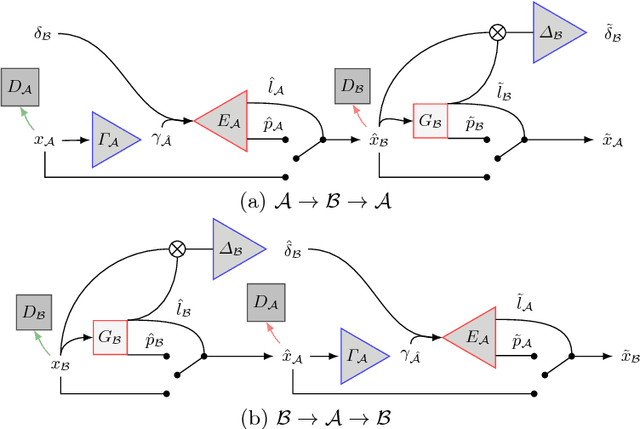
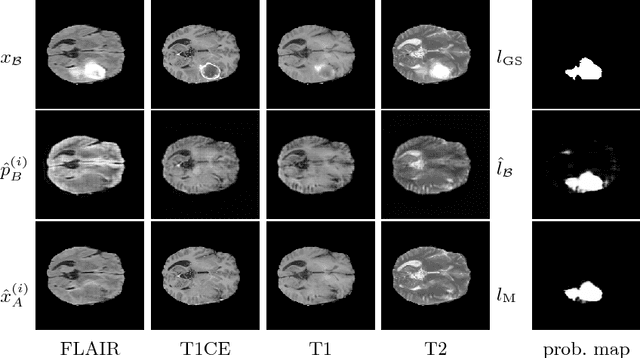
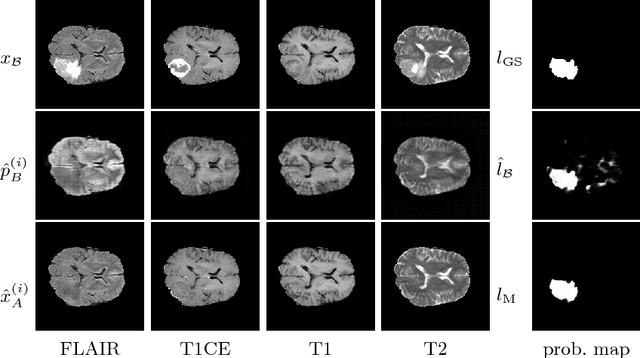
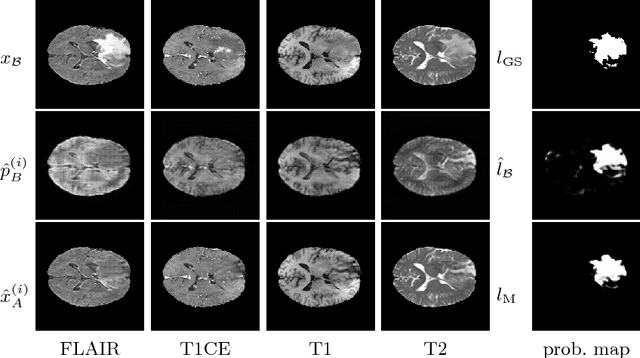
Abstract:We present a method to model pathologies in medical data, trained on data labelled on the image level as healthy or containing a visual defect. Our model not only allows us to create pixelwise semantic segmentations, it is also able to create inpaintings for the segmentations to render the pathological image healthy. Furthermore, we can draw new unseen pathology samples from this model based on the distribution in the data. We show quantitatively, that our method is able to segment pathologies with a surprising accuracy and show qualitative results of both the segmentations and inpaintings. A comparison with a supervised segmentation method indicates, that the accuracy of our proposed weakly-supervised segmentation is nevertheless quite close.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge