Antal Huck
Improved distinct bone segmentation in upper-body CT through multi-resolution networks
Jan 31, 2023Abstract:Purpose: Automated distinct bone segmentation from CT scans is widely used in planning and navigation workflows. U-Net variants are known to provide excellent results in supervised semantic segmentation. However, in distinct bone segmentation from upper body CTs a large field of view and a computationally taxing 3D architecture are required. This leads to low-resolution results lacking detail or localisation errors due to missing spatial context when using high-resolution inputs. Methods: We propose to solve this problem by using end-to-end trainable segmentation networks that combine several 3D U-Nets working at different resolutions. Our approach, which extends and generalizes HookNet and MRN, captures spatial information at a lower resolution and skips the encoded information to the target network, which operates on smaller high-resolution inputs. We evaluated our proposed architecture against single resolution networks and performed an ablation study on information concatenation and the number of context networks. Results: Our proposed best network achieves a median DSC of 0.86 taken over all 125 segmented bone classes and reduces the confusion among similar-looking bones in different locations. These results outperform our previously published 3D U-Net baseline results on the task and distinct-bone segmentation results reported by other groups. Conclusion: The presented multi-resolution 3D U-Nets address current shortcomings in bone segmentation from upper-body CT scans by allowing for capturing a larger field of view while avoiding the cubic growth of the input pixels and intermediate computations that quickly outgrow the computational capacities in 3D. The approach thus improves the accuracy and efficiency of distinct bone segmentation from upper-body CT.
Ensemble uncertainty as a criterion for dataset expansion in distinct bone segmentation from upper-body CT images
Aug 19, 2022
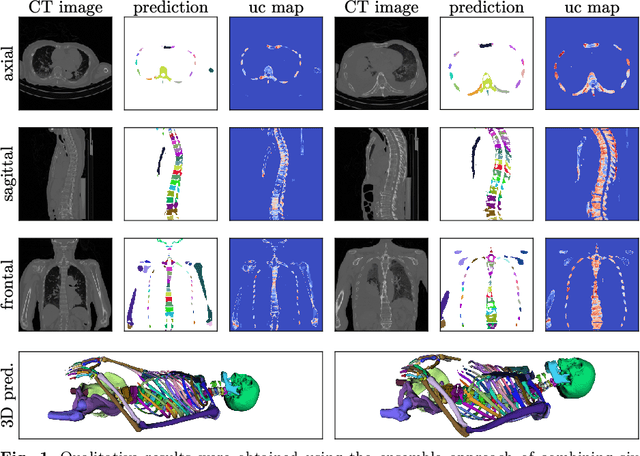
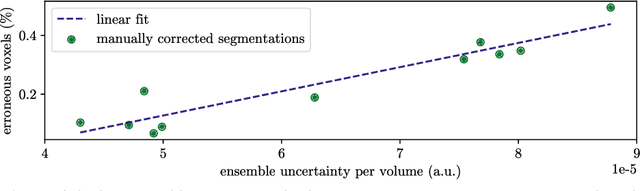
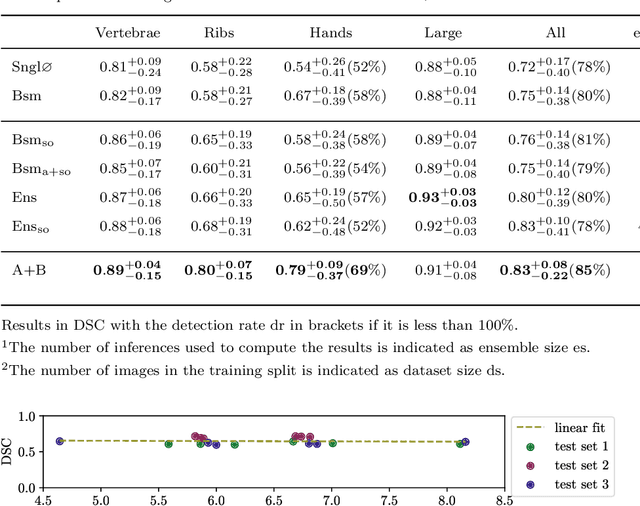
Abstract:Purpose: The localisation and segmentation of individual bones is an important preprocessing step in many planning and navigation applications. It is, however, a time-consuming and repetitive task if done manually. This is true not only for clinical practice but also for the acquisition of training data. We therefore not only present an end-to-end learnt algorithm that is capable of segmenting 125 distinct bones in an upper-body CT, but also provide an ensemble-based uncertainty measure that helps to single out scans to enlarge the training dataset with. Methods We create fully automated end-to-end learnt segmentations using a neural network architecture inspired by the 3D-Unet and fully supervised training. The results are improved using ensembles and inference-time augmentation. We examine the relationship of ensemble-uncertainty to an unlabelled scan's prospective usefulness as part of the training dataset. Results: Our methods are evaluated on an in-house dataset of 16 upper-body CT scans with a resolution of \SI{2}{\milli\meter} per dimension. Taking into account all 125 bones in our label set, our most successful ensemble achieves a median dice score coefficient of 0.83. We find a lack of correlation between a scan's ensemble uncertainty and its prospective influence on the accuracies achieved within an enlarged training set. At the same time, we show that the ensemble uncertainty correlates to the number of voxels that need manual correction after an initial automated segmentation, thus minimising the time required to finalise a new ground truth segmentation. Conclusion: In combination, scans with low ensemble uncertainty need less annotator time while yielding similar future DSC improvements. They are thus ideal candidates to enlarge a training set for upper-body distinct bone segmentation from CT scans. }
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge