Georg Rauter
Robot-mediated physical Human-Human Interaction in Neurorehabilitation: a position paper
Jul 23, 2025



Abstract:Neurorehabilitation conventionally relies on the interaction between a patient and a physical therapist. Robotic systems can improve and enrich the physical feedback provided to patients after neurological injury, but they under-utilize the adaptability and clinical expertise of trained therapists. In this position paper, we advocate for a novel approach that integrates the therapist's clinical expertise and nuanced decision-making with the strength, accuracy, and repeatability of robotics: Robot-mediated physical Human-Human Interaction. This framework, which enables two individuals to physically interact through robotic devices, has been studied across diverse research groups and has recently emerged as a promising link between conventional manual therapy and rehabilitation robotics, harmonizing the strengths of both approaches. This paper presents the rationale of a multidisciplinary team-including engineers, doctors, and physical therapists-for conducting research that utilizes: a unified taxonomy to describe robot-mediated rehabilitation, a framework of interaction based on social psychology, and a technological approach that makes robotic systems seamless facilitators of natural human-human interaction.
Improved distinct bone segmentation in upper-body CT through multi-resolution networks
Jan 31, 2023Abstract:Purpose: Automated distinct bone segmentation from CT scans is widely used in planning and navigation workflows. U-Net variants are known to provide excellent results in supervised semantic segmentation. However, in distinct bone segmentation from upper body CTs a large field of view and a computationally taxing 3D architecture are required. This leads to low-resolution results lacking detail or localisation errors due to missing spatial context when using high-resolution inputs. Methods: We propose to solve this problem by using end-to-end trainable segmentation networks that combine several 3D U-Nets working at different resolutions. Our approach, which extends and generalizes HookNet and MRN, captures spatial information at a lower resolution and skips the encoded information to the target network, which operates on smaller high-resolution inputs. We evaluated our proposed architecture against single resolution networks and performed an ablation study on information concatenation and the number of context networks. Results: Our proposed best network achieves a median DSC of 0.86 taken over all 125 segmented bone classes and reduces the confusion among similar-looking bones in different locations. These results outperform our previously published 3D U-Net baseline results on the task and distinct-bone segmentation results reported by other groups. Conclusion: The presented multi-resolution 3D U-Nets address current shortcomings in bone segmentation from upper-body CT scans by allowing for capturing a larger field of view while avoiding the cubic growth of the input pixels and intermediate computations that quickly outgrow the computational capacities in 3D. The approach thus improves the accuracy and efficiency of distinct bone segmentation from upper-body CT.
Using Supervised Deep-Learning to Model Edge-FBG Shape Sensors
Oct 28, 2022Abstract:Continuum robots in robot-assisted minimally invasive surgeries provide adequate access to target anatomies that are not directly reachable through small incisions. Achieving precise and reliable motion control of such snake-like manipulators necessitates an accurate navigation system that requires no line-of-sight and is immune to electromagnetic noises. Fiber Bragg Grating (FBG) shape sensors, particularly edge-FBGs, are promising tools for this task. However, in edge-FBG sensors, the intensity ratio between Bragg wavelengths carries the strain information that can be affected by undesired bending-related phenomena, making standard characterization techniques less suitable for these sensors. We showed in our previous work that a deep learning model has the potential to extract the strain information from the full edge-FBG spectrum and accurately predict the sensor's shape. In this paper, we conduct a more thorough investigation to find a suitable architectural design with lower prediction errors. We use the Hyperband algorithm to search for optimal hyperparameters in two steps. First, we limit the search space to layer settings, where the best-performing configuration gets selected. Then, we modify the search space for tuning the training and loss calculation hyperparameters. We also analyze various data transformations on the input and output variables, as data rescaling can directly influence the model's performance. Moreover, we performed discriminative training using Siamese network architecture that employs two CNNs with identical parameters to learn similarity metrics between the spectra of similar target values. The best-performing network architecture among all evaluated configurations can predict the sensor's shape with a median tip error of 3.11 mm.
The secret role of undesired physical effects in accurate shape sensing with eccentric FBGs
Oct 28, 2022Abstract:Fiber optic shape sensors have enabled unique advances in various navigation tasks, from medical tool tracking to industrial applications. Eccentric fiber Bragg gratings (FBG) are cheap and easy-to-fabricate shape sensors that are often interrogated with simple setups. However, using low-cost interrogation systems for such intensity-based quasi-distributed sensors introduces further complications to the sensor's signal. Therefore, eccentric FBGs have not been able to accurately estimate complex multi-bend shapes. Here, we present a novel technique to overcome these limitations and provide accurate and precise shape estimation in eccentric FBG sensors. We investigate the most important bending-induced effects in curved optical fibers that are usually eliminated in intensity-based fiber sensors. These effects contain shape deformation information with a higher spatial resolution that we are now able to extract using deep learning techniques. We design a deep learning model based on a convolutional neural network that is trained to predict shapes given the sensor's spectra. We also provide a visual explanation, highlighting wavelength elements whose intensities are more relevant in making shape predictions. These findings imply that deep learning techniques benefit from the bending-induced effects that impact the desired signal in a complex manner. This is the first step toward cheap yet accurate fiber shape sensing solutions.
Ensemble uncertainty as a criterion for dataset expansion in distinct bone segmentation from upper-body CT images
Aug 19, 2022
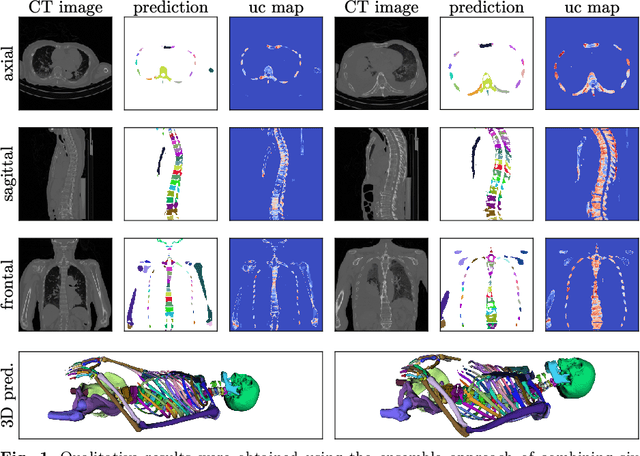
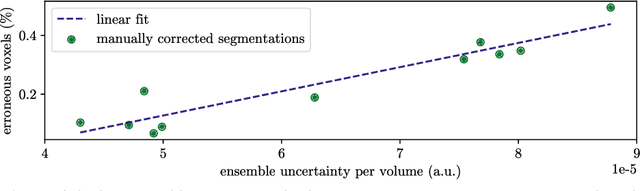
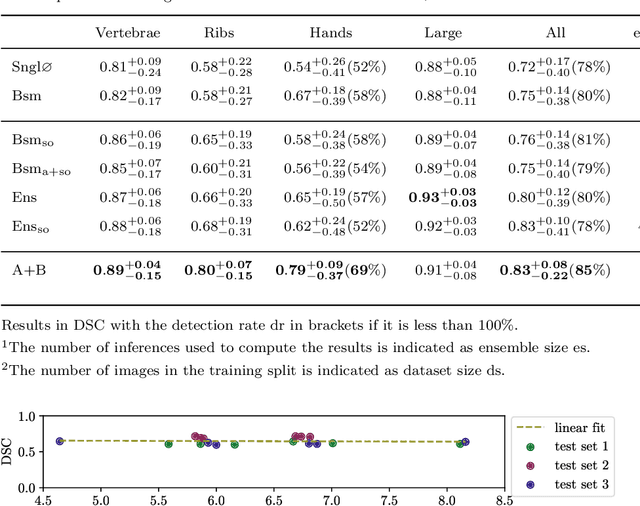
Abstract:Purpose: The localisation and segmentation of individual bones is an important preprocessing step in many planning and navigation applications. It is, however, a time-consuming and repetitive task if done manually. This is true not only for clinical practice but also for the acquisition of training data. We therefore not only present an end-to-end learnt algorithm that is capable of segmenting 125 distinct bones in an upper-body CT, but also provide an ensemble-based uncertainty measure that helps to single out scans to enlarge the training dataset with. Methods We create fully automated end-to-end learnt segmentations using a neural network architecture inspired by the 3D-Unet and fully supervised training. The results are improved using ensembles and inference-time augmentation. We examine the relationship of ensemble-uncertainty to an unlabelled scan's prospective usefulness as part of the training dataset. Results: Our methods are evaluated on an in-house dataset of 16 upper-body CT scans with a resolution of \SI{2}{\milli\meter} per dimension. Taking into account all 125 bones in our label set, our most successful ensemble achieves a median dice score coefficient of 0.83. We find a lack of correlation between a scan's ensemble uncertainty and its prospective influence on the accuracies achieved within an enlarged training set. At the same time, we show that the ensemble uncertainty correlates to the number of voxels that need manual correction after an initial automated segmentation, thus minimising the time required to finalise a new ground truth segmentation. Conclusion: In combination, scans with low ensemble uncertainty need less annotator time while yielding similar future DSC improvements. They are thus ideal candidates to enlarge a training set for upper-body distinct bone segmentation from CT scans. }
3D Segmentation Networks for Excessive Numbers of Classes: Distinct Bone Segmentation in Upper Bodies
Oct 14, 2020



Abstract:Segmentation of distinct bones plays a crucial role in diagnosis, planning, navigation, and the assessment of bone metastasis. It supplies semantic knowledge to visualisation tools for the planning of surgical interventions and the education of health professionals. Fully supervised segmentation of 3D data using Deep Learning methods has been extensively studied for many tasks but is usually restricted to distinguishing only a handful of classes. With 125 distinct bones, our case includes many more labels than typical 3D segmentation tasks. For this reason, the direct adaptation of most established methods is not possible. This paper discusses the intricacies of training a 3D segmentation network in a many-label setting and shows necessary modifications in network architecture, loss function, and data augmentation. As a result, we demonstrate the robustness of our method by automatically segmenting over one hundred distinct bones simultaneously in an end-to-end learnt fashion from a CT-scan.
* 10 pages, 3 figures, 2 tables, accepted into MICCAI 2020 International Workshop on Machine Learning in Medical Imaging
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge