Mufan Xu
Instruction Anchors: Dissecting the Causal Dynamics of Modality Arbitration
Feb 03, 2026Abstract:Modality following serves as the capacity of multimodal large language models (MLLMs) to selectively utilize multimodal contexts based on user instructions. It is fundamental to ensuring safety and reliability in real-world deployments. However, the underlying mechanisms governing this decision-making process remain poorly understood. In this paper, we investigate its working mechanism through an information flow lens. Our findings reveal that instruction tokens function as structural anchors for modality arbitration: Shallow attention layers perform non-selective information transfer, routing multimodal cues to these anchors as a latent buffer; Modality competition is resolved within deep attention layers guided by the instruction intent, while MLP layers exhibit semantic inertia, acting as an adversarial force. Furthermore, we identify a sparse set of specialized attention heads that drive this arbitration. Causal interventions demonstrate that manipulating a mere $5\%$ of these critical heads can decrease the modality-following ratio by $60\%$ through blocking, or increase it by $60\%$ through targeted amplification of failed samples. Our work provides a substantial step toward model transparency and offers a principled framework for the orchestration of multimodal information in MLLMs.
Memory-augmented Query Reconstruction for LLM-based Knowledge Graph Reasoning
Mar 07, 2025



Abstract:Large language models (LLMs) have achieved remarkable performance on knowledge graph question answering (KGQA) tasks by planning and interacting with knowledge graphs. However, existing methods often confuse tool utilization with knowledge reasoning, harming readability of model outputs and giving rise to hallucinatory tool invocations, which hinder the advancement of KGQA. To address this issue, we propose Memory-augmented Query Reconstruction for LLM-based Knowledge Graph Reasoning (MemQ) to decouple LLM from tool invocation tasks using LLM-built query memory. By establishing a memory module with explicit descriptions of query statements, the proposed MemQ facilitates the KGQA process with natural language reasoning and memory-augmented query reconstruction. Meanwhile, we design an effective and readable reasoning to enhance the LLM's reasoning capability in KGQA. Experimental results that MemQ achieves state-of-the-art performance on widely used benchmarks WebQSP and CWQ.
LLM-based Discriminative Reasoning for Knowledge Graph Question Answering
Dec 17, 2024



Abstract:Large language models (LLMs) based on generative pre-trained Transformer have achieved remarkable performance on knowledge graph question-answering (KGQA) tasks. However, LLMs often produce ungrounded subgraph planning or reasoning results in KGQA due to the hallucinatory behavior brought by the generative paradigm, which may hinder the advancement of the LLM-based KGQA model. To deal with the issue, we propose a novel LLM-based Discriminative Reasoning (LDR) method to explicitly model the subgraph retrieval and answer inference process. By adopting discriminative strategies, the proposed LDR method not only enhances the capability of LLMs to retrieve question-related subgraphs but also alleviates the issue of ungrounded reasoning brought by the generative paradigm of LLMs. Experimental results show that the proposed approach outperforms multiple strong comparison methods, along with achieving state-of-the-art performance on two widely used WebQSP and CWQ benchmarks.
Impact-Oriented Contextual Scholar Profiling using Self-Citation Graphs
Apr 24, 2023
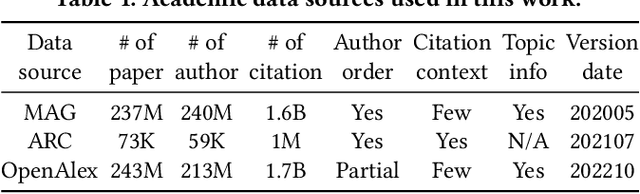
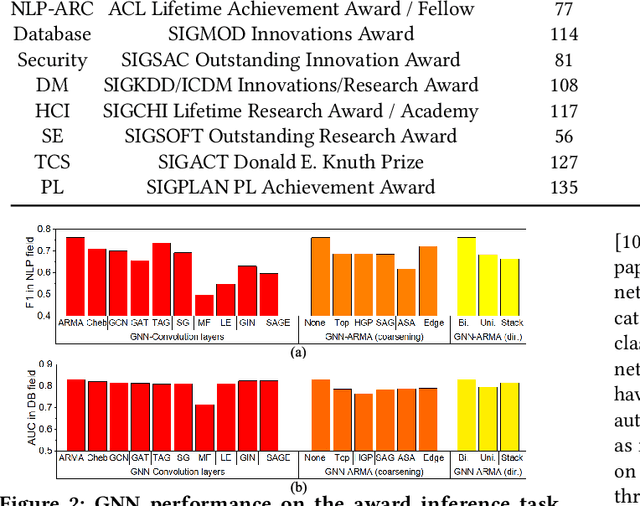
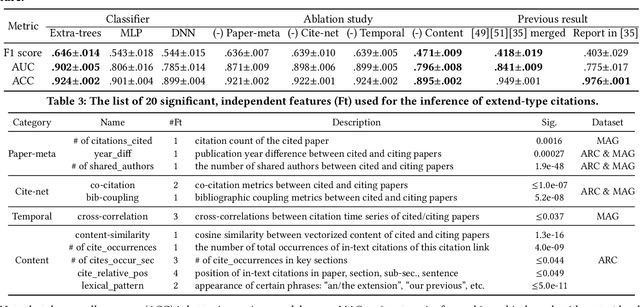
Abstract:Quantitatively profiling a scholar's scientific impact is important to modern research society. Current practices with bibliometric indicators (e.g., h-index), lists, and networks perform well at scholar ranking, but do not provide structured context for scholar-centric, analytical tasks such as profile reasoning and understanding. This work presents GeneticFlow (GF), a suite of novel graph-based scholar profiles that fulfill three essential requirements: structured-context, scholar-centric, and evolution-rich. We propose a framework to compute GF over large-scale academic data sources with millions of scholars. The framework encompasses a new unsupervised advisor-advisee detection algorithm, a well-engineered citation type classifier using interpretable features, and a fine-tuned graph neural network (GNN) model. Evaluations are conducted on the real-world task of scientific award inference. Experiment outcomes show that the F1 score of best GF profile significantly outperforms alternative methods of impact indicators and bibliometric networks in all the 6 computer science fields considered. Moreover, the core GF profiles, with 63.6%-66.5% nodes and 12.5%-29.9% edges of the full profile, still significantly outrun existing methods in 5 out of 6 fields studied. Visualization of GF profiling result also reveals human explainable patterns for high-impact scholars.
Inference with Hybrid Bio-hardware Neural Networks
May 28, 2019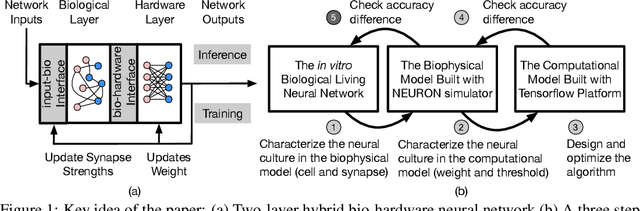
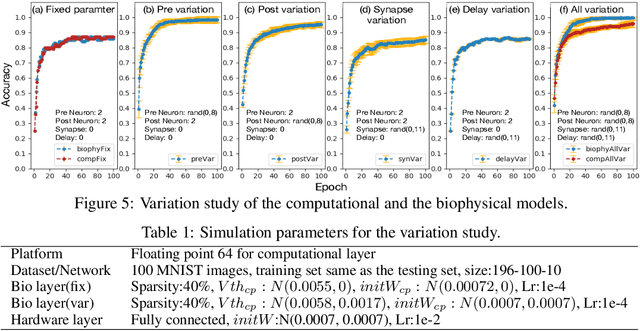
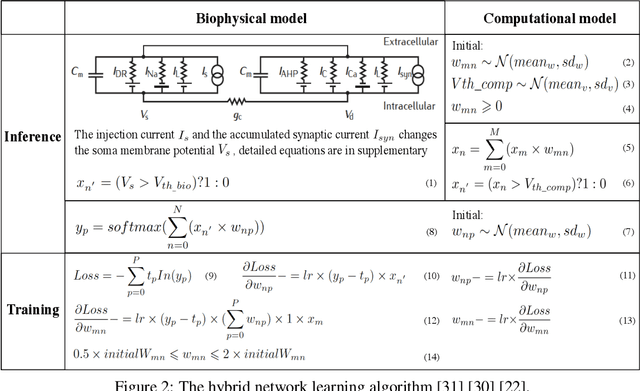
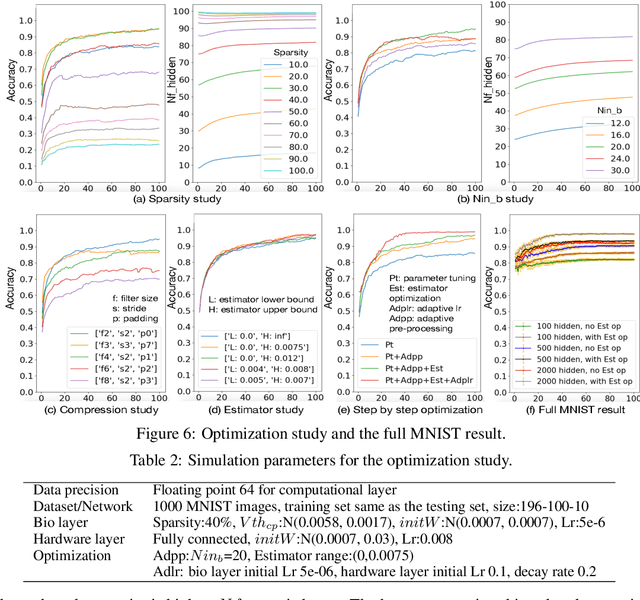
Abstract:To understand the learning process in brains, biologically plausible algorithms have been explored by modeling the detailed neuron properties and dynamics. On the other hand, simplified multi-layer models of neural networks have shown great success on computational tasks such as image classification and speech recognition. However, the computational models that can achieve good accuracy for these learning applications are very different from the bio-plausible models. This paper studies whether a bio-plausible model of a in vitro living neural network can be used to perform machine learning tasks and achieve good inference accuracy. A novel two-layer bio-hardware hybrid neural network is proposed. The biological layer faithfully models variations of synapses, neurons, and network sparsity in in vitro living neural networks. The hardware layer is a computational fully-connected layer that tunes parameters to optimize for accuracy. Several techniques are proposed to improve the inference accuracy of the proposed hybrid neural network. For instance, an adaptive pre-processing technique helps the proposed neural network to achieve good learning accuracy for different living neural network sparsity. The proposed hybrid neural network with realistic neuron parameters and variations achieves a 98.3% testing accuracy for the handwritten digit recognition task on the full MNIST dataset.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge