Mohammed Baharoon
RadGame: An AI-Powered Platform for Radiology Education
Sep 16, 2025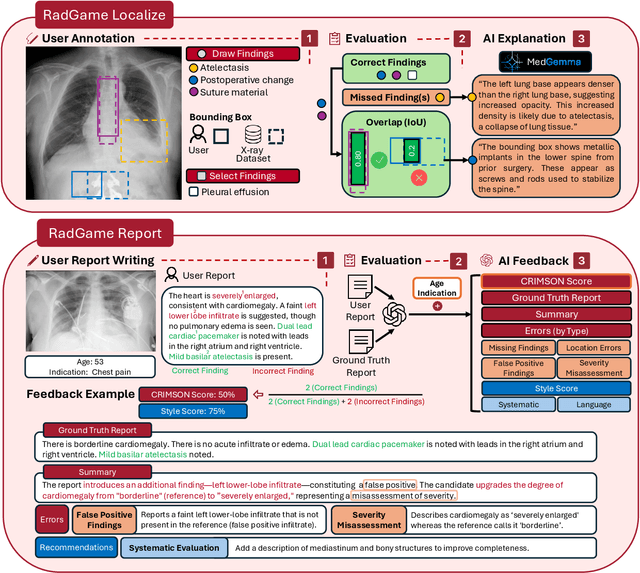
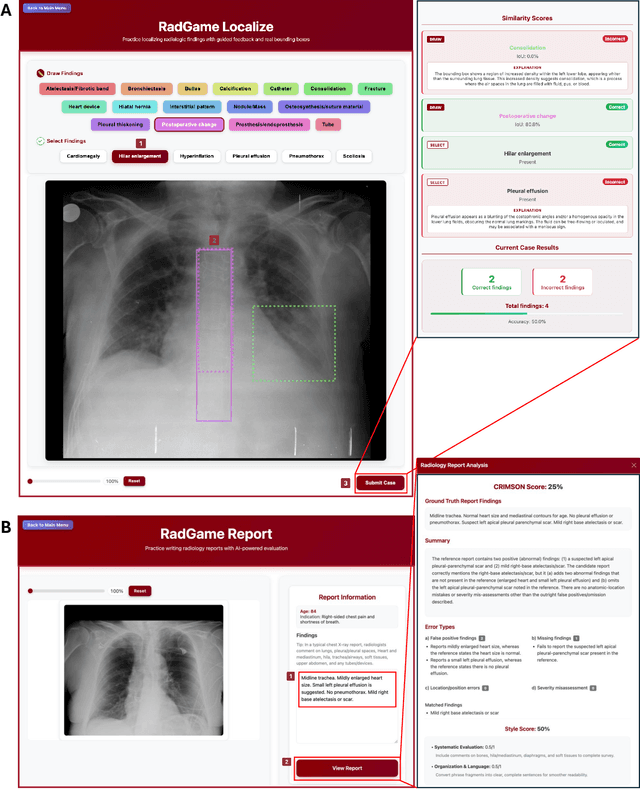
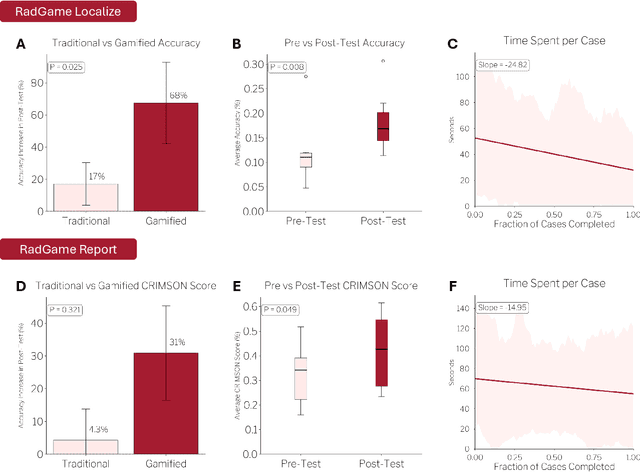
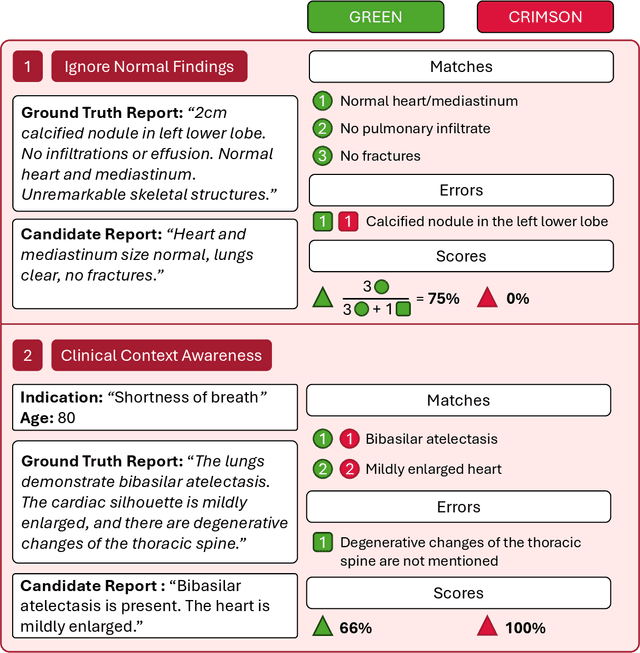
Abstract:We introduce RadGame, an AI-powered gamified platform for radiology education that targets two core skills: localizing findings and generating reports. Traditional radiology training is based on passive exposure to cases or active practice with real-time input from supervising radiologists, limiting opportunities for immediate and scalable feedback. RadGame addresses this gap by combining gamification with large-scale public datasets and automated, AI-driven feedback that provides clear, structured guidance to human learners. In RadGame Localize, players draw bounding boxes around abnormalities, which are automatically compared to radiologist-drawn annotations from public datasets, and visual explanations are generated by vision-language models for user missed findings. In RadGame Report, players compose findings given a chest X-ray, patient age and indication, and receive structured AI feedback based on radiology report generation metrics, highlighting errors and omissions compared to a radiologist's written ground truth report from public datasets, producing a final performance and style score. In a prospective evaluation, participants using RadGame achieved a 68% improvement in localization accuracy compared to 17% with traditional passive methods and a 31% improvement in report-writing accuracy compared to 4% with traditional methods after seeing the same cases. RadGame highlights the potential of AI-driven gamification to deliver scalable, feedback-rich radiology training and reimagines the application of medical AI resources in education.
ReXGroundingCT: A 3D Chest CT Dataset for Segmentation of Findings from Free-Text Reports
Jul 29, 2025Abstract:We present ReXGroundingCT, the first publicly available dataset to link free-text radiology findings with pixel-level segmentations in 3D chest CT scans that is manually annotated. While prior datasets have relied on structured labels or predefined categories, ReXGroundingCT captures the full expressiveness of clinical language represented in free text and grounds it to spatially localized 3D segmentation annotations in volumetric imaging. This addresses a critical gap in medical AI: the ability to connect complex, descriptive text, such as "3 mm nodule in the left lower lobe", to its precise anatomical location in three-dimensional space, a capability essential for grounded radiology report generation systems. The dataset comprises 3,142 non-contrast chest CT scans paired with standardized radiology reports from the CT-RATE dataset. Using a systematic three-stage pipeline, GPT-4 was used to extract positive lung and pleural findings, which were then manually segmented by expert annotators. A total of 8,028 findings across 16,301 entities were annotated, with quality control performed by board-certified radiologists. Approximately 79% of findings are focal abnormalities, while 21% are non-focal. The training set includes up to three representative segmentations per finding, while the validation and test sets contain exhaustive labels for each finding entity. ReXGroundingCT establishes a new benchmark for developing and evaluating sentence-level grounding and free-text medical segmentation models in chest CT. The dataset can be accessed at https://huggingface.co/datasets/rajpurkarlab/ReXGroundingCT.
Exploring the Design Space of 3D MLLMs for CT Report Generation
Jun 26, 2025Abstract:Multimodal Large Language Models (MLLMs) have emerged as a promising way to automate Radiology Report Generation (RRG). In this work, we systematically investigate the design space of 3D MLLMs, including visual input representation, projectors, Large Language Models (LLMs), and fine-tuning techniques for 3D CT report generation. We also introduce two knowledge-based report augmentation methods that improve performance on the GREEN score by up to 10\%, achieving the 2nd place on the MICCAI 2024 AMOS-MM challenge. Our results on the 1,687 cases from the AMOS-MM dataset show that RRG is largely independent of the size of LLM under the same training protocol. We also show that larger volume size does not always improve performance if the original ViT was pre-trained on a smaller volume size. Lastly, we show that using a segmentation mask along with the CT volume improves performance. The code is publicly available at https://github.com/bowang-lab/AMOS-MM-Solution
MedSAM2: Segment Anything in 3D Medical Images and Videos
Apr 04, 2025Abstract:Medical image and video segmentation is a critical task for precision medicine, which has witnessed considerable progress in developing task or modality-specific and generalist models for 2D images. However, there have been limited studies on building general-purpose models for 3D images and videos with comprehensive user studies. Here, we present MedSAM2, a promptable segmentation foundation model for 3D image and video segmentation. The model is developed by fine-tuning the Segment Anything Model 2 on a large medical dataset with over 455,000 3D image-mask pairs and 76,000 frames, outperforming previous models across a wide range of organs, lesions, and imaging modalities. Furthermore, we implement a human-in-the-loop pipeline to facilitate the creation of large-scale datasets resulting in, to the best of our knowledge, the most extensive user study to date, involving the annotation of 5,000 CT lesions, 3,984 liver MRI lesions, and 251,550 echocardiogram video frames, demonstrating that MedSAM2 can reduce manual costs by more than 85%. MedSAM2 is also integrated into widely used platforms with user-friendly interfaces for local and cloud deployment, making it a practical tool for supporting efficient, scalable, and high-quality segmentation in both research and healthcare environments.
Segment Anything in Medical Images and Videos: Benchmark and Deployment
Aug 06, 2024



Abstract:Recent advances in segmentation foundation models have enabled accurate and efficient segmentation across a wide range of natural images and videos, but their utility to medical data remains unclear. In this work, we first present a comprehensive benchmarking of the Segment Anything Model 2 (SAM2) across 11 medical image modalities and videos and point out its strengths and weaknesses by comparing it to SAM1 and MedSAM. Then, we develop a transfer learning pipeline and demonstrate SAM2 can be quickly adapted to medical domain by fine-tuning. Furthermore, we implement SAM2 as a 3D slicer plugin and Gradio API for efficient 3D image and video segmentation. The code has been made publicly available at \url{https://github.com/bowang-lab/MedSAM}.
Harmony: A Joint Self-Supervised and Weakly-Supervised Framework for Learning General Purpose Visual Representations
May 23, 2024Abstract:Vision-language contrastive learning frameworks like CLIP enable learning representations from natural language supervision, and provide strong zero-shot classification capabilities. However, due to the nature of the supervisory signal in these paradigms, they lack the ability to learn localized features, leading to degraded performance on dense prediction tasks like segmentation and detection. On the other hand, self-supervised learning methods have shown the ability to learn granular representations, complementing the high-level features in vision-language training. In this work, we present Harmony, a framework that combines vision-language training with discriminative and generative self-supervision to learn visual features that can be generalized across vision downstream tasks. Our framework is specifically designed to work on web-scraped data by not relying on negative examples and addressing the one-to-one correspondence issue using soft CLIP targets generated by an EMA model. We comprehensively evaluate Harmony across various vision downstream tasks and find that it significantly outperforms the baseline CLIP and the previously leading joint self and weakly-supervised methods, MaskCLIP and SLIP. Specifically, when comparing against these methods, Harmony shows superior performance in fine-tuning and zero-shot classification on ImageNet-1k, semantic segmentation on ADE20K, and both object detection and instance segmentation on MS-COCO, when pre-training a ViT-S/16 on CC3M. We also show that Harmony outperforms other self-supervised learning methods like iBOT and MAE across all tasks evaluated. On https://github.com/MohammedSB/Harmony our code is publicly available.
Towards General Purpose Vision Foundation Models for Medical Image Analysis: An Experimental Study of DINOv2 on Radiology Benchmarks
Dec 07, 2023



Abstract:The integration of deep learning systems into the medical domain has been hindered by the resource-intensive process of data annotation and the inability of these systems to generalize to different data distributions. Foundation models, which are models pre-trained on large datasets, have emerged as a solution to reduce reliance on annotated data and enhance model generalizability and robustness. DINOv2, an open-source foundation model pre-trained with self-supervised learning on 142 million curated natural images, excels in extracting general-purpose visual representations, exhibiting promising capabilities across various vision tasks. Nevertheless, a critical question remains unanswered regarding DINOv2's adaptability to radiological imaging, and the clarity on whether its features are sufficiently general to benefit radiology image analysis is yet to be established. Therefore, this study comprehensively evaluates DINOv2 for radiology, conducting over 100 experiments across diverse modalities (X-ray, CT, and MRI). Tasks include disease classification and organ segmentation on both 2D and 3D images, evaluated under different settings like kNN, few-shot learning, linear-probing, end-to-end fine-tuning, and parameter-efficient fine-tuning, to measure the effectiveness and generalizability of the DINOv2 feature embeddings. Comparative analyses with established medical image analysis models, U-Net and TransUnet for segmentation, and CNN and ViT models pre-trained via supervised, weakly supervised, and self-supervised learning for classification, reveal DINOv2's superior performance in segmentation tasks and competitive results in disease classification. The findings contribute insights to potential avenues for optimizing pre-training strategies for medical imaging and enhancing the broader understanding of DINOv2's role in bridging the gap between natural and radiological image analysis.
HyMNet: a Multimodal Deep Learning System for Hypertension Classification using Fundus Photographs and Cardiometabolic Risk Factors
Oct 02, 2023Abstract:In recent years, deep learning has shown promise in predicting hypertension (HTN) from fundus images. However, most prior research has primarily focused on analyzing a single type of data, which may not capture the full complexity of HTN risk. To address this limitation, this study introduces a multimodal deep learning (MMDL) system, dubbed HyMNet, which combines fundus images and cardiometabolic risk factors, specifically age and gender, to improve hypertension detection capabilities. Our MMDL system uses the DenseNet-201 architecture, pre-trained on ImageNet, for the fundus imaging path and a fully connected neural network for the age and gender path. The two paths are jointly trained by concatenating 64 features output from each path that are then fed into a fusion network. The system was trained on 1,143 retinal images from 626 individuals collected from the Saudi Ministry of National Guard Health Affairs. The results show that the multimodal model that integrates fundus images along with age and gender achieved an AUC of 0.791 [CI: 0.735, 0.848], which outperforms the unimodal model trained solely on fundus photographs that yielded an AUC of 0.766 [CI: 0.705, 0.828] for hypertension detection.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge